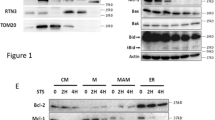
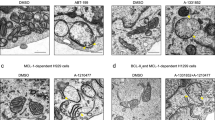
Abstract
MCL-1, an anti-apoptotic BCL-2 family member that is essential for the survival of multiple cell lineages, is also among the most highly amplified genes in cancer. Although MCL-1 is known to oppose cell death, precisely how it functions to promote survival of normal and malignant cells is poorly understood. Here, we report that different forms of MCL-1 reside in distinct mitochondrial locations and exhibit separable functions. On the outer mitochondrial membrane, an MCL-1 isoform acts like other anti-apoptotic BCL-2 molecules to antagonize apoptosis, whereas an amino-terminally truncated isoform of MCL-1 that is imported into the mitochondrial matrix is necessary to facilitate normal mitochondrial fusion, ATP production, membrane potential, respiration, cristae ultrastructure and maintenance of oligomeric ATP synthase. Our results provide insight into how the surprisingly diverse salutary functions of MCL-1 may control the survival of both normal and cancer cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
17 May 2012
In the version of this article initially published online, the x axes for Fig. 4b,c were incorrectly labelled. The correct units are μM. This has been corrected.
References
Danial, N. N. & Korsmeyer, S. J. Cell death: critical control points. Cell 116, 205–219 (2004).
Cory, S. & Adams, J. M. The Bcl2 family: regulators of the cellular life-or-death switch. Nat. Rev. Cancer 2, 647–656 (2002).
Youle, R. J. & Strasser, A. The BCL-2 protein family: opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 9, 47–59 (2008).
Wei, M. C. et al. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science 292, 727–730 (2001).
Goldstein, J. C., Waterhouse, N. J., Juin, P., Evan, G. I. & Green, D. R. The coordinate release of cytochrome c during apoptosis is rapid, complete and kinetically invariant. Nat. Cell Biol. 2, 156–162 (2000).
Cheng, E. H. et al. BCL-2, BCL-X(L) sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol. Cell 8, 705–711 (2001).
Zong, W. X., Lindsten, T., Ross, A. J., MacGregor, G. R. & Thompson, C. B. BH3-only proteins that bind pro-survival Bcl-2 family members fail to induce apoptosis in the absence of Bax and Bak. Genes Dev. 15, 1481–1486 (2001).
Frey, T. G. & Mannella, C. A. The internal structure of mitochondria. Trends Biochem. Sci. 25, 319–324 (2000).
Chan, D. C. Mitochondrial dynamics in disease. New Engl. J. Med. 356, 1707–1709 (2007).
Chen, H. & Chan, D. C. Emerging functions of mammalian mitochondrial fusion and fission. Hum. Mol. Genet. 14 (Spec no. 2), R283–R289 (2005).
Mitra, K., Wunder, C., Roysam, B., Lin, G. & Lippincott-Schwartz, J. A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc. Natl Acad. Sci. USA 106, 11960–11965 (2009).
Chen, H. et al. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 160, 189–200 (2003).
Bleazard, W. et al. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1, 298–304 (1999).
Rinkenberger, J. L., Horning, S., Klocke, B., Roth, K. & Korsmeyer, S. J. Mcl-1 deficiency results in peri-implantation embryonic lethality. Genes Dev. 14, 23–27 (2000).
Opferman, J. T. et al. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature 426, 671–676 (2003).
Dzhagalov, I., Dunkle, A. & He, Y. W. The anti-apoptotic Bcl-2 family member Mcl-1 promotes T lymphocyte survival at multiple stages. J. Immunol. 181, 521–528 (2008).
Opferman, J. T. et al. Obligate role of anti-apoptotic MCL-1 in the survival of hematopoietic stem cells. Science 307, 1101–1104 (2005).
Dzhagalov, I., St John, A. & He, Y. W. The antiapoptotic protein Mcl-1 is essential for the survival of neutrophils but not macrophages. Blood 109, 1620–1626 (2007).
Steimer, D. A. et al. Selective roles for antiapoptotic MCL-1 during granulocyte development and macrophage effector function. Blood 113, 2805–2815 (2009).
Arbour, N. et al. Mcl-1 is a key regulator of apoptosis during CNS development and after DNA damage. J. Neurosci. 28, 6068–6078 (2008).
Beroukhim, R. et al. The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010).
Wei, G. et al. Gene expression-based chemical genomics identifies rapamycin as a modulator of MCL1 and glucocorticoid resistance. Cancer Cell 10, 331–342 (2006).
Wuilleme-Toumi, S. et al. Mcl-1 is overexpressed in multiple myeloma and associated with relapse and shorter survival. Leukemia 19, 1248–1252 (2005).
Vogel, F., Bornhovd, C., Neupert, W. & Reichert, A. S. Dynamic subcompartmentalization of the mitochondrial inner membrane. J. Cell Biol. 175, 237–247 (2006).
Gilkerson, R. W., Selker, J. M. & Capaldi, R. A. The cristal membrane of mitochondria is the principal site of oxidative phosphorylation. FEBS Lett. 546, 355–358 (2003).
Maurer, U., Charvet, C., Wagman, A. S., Dejardin, E. & Green, D. R. Glycogen synthase kinase-3 regulates mitochondrial outer membrane permeabilization and apoptosis by destabilization of MCL-1. Mol. Cell 21, 749–760 (2006).
Domina, A. M., Vrana, J. A., Gregory, M. A., Hann, S. R. & Craig, R. W. MCL1 is phosphorylated in the PEST region and stabilized upon ERK activation in viable cells, and at additional sites with cytotoxic okadaic acid or taxol. Oncogene 23, 5301–5315 (2004).
Morel, C., Carlson, S. M., White, F. M. & Davis, R. J. Mcl-1 integrates the opposing actions of signaling pathways that mediate survival and apoptosis. Mol. Cell Biol. 29, 3845–3852 (2009).
Warr, M. R. & Shore, G. C. Unique biology of Mcl-1: therapeutic opportunities in cancer. Curr. Mol. Med. 8, 138–147 (2008).
Kojima, S., Hyakutake, A., Koshikawa, N., Nakagawara, A. & Takenaga, K. MCL-1V, a novel mouse antiapoptotic MCL-1 variant, generated by RNA splicing at a non-canonical splicing pair. Biochem. Biophys. Res. Commun. 391, 492–497 (2010).
De Biasio, A. et al. N-terminal truncation of antiapoptotic MCL1, but not G2/M-induced phosphorylation, is associated with stabilization and abundant expression in tumor cells. J. Biol. Chem. 282, 23919–23936 (2007).
Huang, C. R. & Yang-Yen, H. F. The fast-mobility isoform of mouse Mcl-1 is a mitochondrial matrix-localized protein with attenuated anti-apoptotic activity. FEBS Lett. 584, 3323–3330 (2010).
Schmidt, O., Pfanner, N. & Meisinger, C. Mitochondrial protein import: from proteomics to functional mechanisms. Nat. Rev. Mol. Cell Biol. 11, 655–667 (2010).
Pfanner, N., Muller, H. K., Harmey, M. A. & Neupert, W. Mitochondrial protein import: involvement of the mature part of a cleavable precursor protein in the binding to receptor sites. EMBO J. 6, 3449–3454 (1987).
Karbowski, M. et al. Quantitation of mitochondrial dynamics by photolabeling of individual organelles shows that mitochondrial fusion is blocked during the Bax activation phase of apoptosis. J. Cell Biol. 164, 493–499 (2004).
Wallace, D. C. Mitochondrial diseases in man and mouse. Science 283, 1482–1488 (1999).
Reitzer, L. J., Wice, B. M. & Kennell, D. Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J. Biol. Chem. 254, 2669–2676 (1979).
Rossignol, R. et al. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res. 64, 985–993 (2004).
Ferrick, D. A., Neilson, A. & Beeson, C. Advances in measuring cellular bioenergetics using extracellular flux. Drug Discov. Today 13, 268–274 (2008).
Acin-Perez, R., Fernandez-Silva, P., Peleato, M. L., Perez-Martos, A. & Enriquez, J. A. Respiratory active mitochondrial supercomplexes. Mol. Cell 32, 529–539 (2008).
Genova, M. L. et al. Is supercomplex organization of the respiratory chain required for optimal electron transfer activity? Biochim. Biophys. Acta 1777, 740–746 (2008).
Diaz, F., Fukui, H., Garcia, S. & Moraes, C. T. Cytochrome c oxidase is required for the assembly/stability of respiratory complex I in mouse fibroblasts. Mol. Cell Biol. 26, 4872–4881 (2006).
Krause, F., Reifschneider, N. H., Goto, S. & Dencher, N. A. Active oligomeric ATP synthases in mammalian mitochondria. Biochem. Biophys. Res. Commun. 329, 583–590 (2005).
Giraud, M. F. et al. Is there a relationship between the supramolecular organization of the mitochondrial ATP synthase and the formation of cristae? Biochim. Biophys. Acta 1555, 174–180 (2002).
Thomas, D. et al. Supramolecular organization of the yeast F1Fo-ATP synthase. Biol. Cell 100, 591–601 (2008).
Gomes, L. C., Di Benedetto, G. & Scorrano, L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 13, 589–598 (2011).
Paumard, P. et al. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 21, 221–230 (2002).
Bornhovd, C., Vogel, F., Neupert, W. & Reichert, A. S. Mitochondrial membrane potential is dependent on the oligomeric state of F1F0-ATP synthase supracomplexes. J. Biol. Chem. 281, 13990–13998 (2006).
Kozopas, K. M., Yang, T., Buchan, H. L., Zhou, P. & Craig, R. W. MCL1, a gene expressed in programmed myeloid cell differentiation, has sequence similarity to BCL2. Proc. Natl Acad. Sci. USA 90, 3516–3520 (1993).
Krajewski, S. et al. Immunohistochemical analysis of Mcl-1 protein in human tissues. Differential regulation of Mcl-1 and Bcl-2 protein production suggests a unique role for Mcl-1 in control of programmed cell death in vivo. Am. J. Pathol. 146, 1309–1319 (1995).
Yang, T., Kozopas, K. M. & Craig, R. W. The intracellular distribution and pattern of expression of Mcl-1 overlap with, but are not identical to, those of Bcl-2. J. Cell Biol. 128, 1173–1184 (1995).
Chen, H. et al. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell 141, 280–289 (2010).
Velours, J., Dautant, A., Salin, B., Sagot, I. & Brethes, D. Mitochondrial F1F0-ATP synthase and organellar internal architecture. Int. J. Biochem. Cell Biol. 41, 1783–1789 (2009).
Deberardinis, R. J., Sayed, N., Ditsworth, D. & Thompson, C. B. Brick by brick: metabolism and tumor cell growth. Curr. Opin. Genet. Dev. 18, 54–61 (2008).
Stewart, D. P. et al. Ubiquitin-independent degradation of antiapoptotic MCL-1. Mol. Cell Biol. 30, 3099–3110 (2010).
Koopman, W. J. et al. Inhibition of complex I of the electron transport chain causes O2−.-mediated mitochondrial outgrowth. Am. J. Physiol. Cell Physiol. 288, C1440–C1450 (2005).
Peters, P. J., Bos, E. & Griekspoor, A. Cryo-immunogold electron microscopy. Curr. Protoc. Cell. Biol. Chapter 4 (2006) Unit 4 7.
Frost, M. T., Wang, Q., Moncada, S. & Singer, M. Hypoxia accelerates nitric oxide-dependent inhibition of mitochondrial complex I in activated macrophages. Am. J. Physiol. Regul. Integr. Comp. Physiol. 288, R394–R400 (2005).
Miyadera, H. et al. Atpenins, potent and specific inhibitors of mitochondrial complex II (succinate-ubiquinone oxidoreductase). Proc. Natl Acad. Sci. USA 100, 473–477 (2003).
Miro, O. et al. Cytochrome c oxidase assay in minute amounts of human skeletal muscle using single wavelength spectrophotometers. J. Neurosci. Methods 80, 107–111 (1998).
Hinman, L. M. & Blass, J. P. An NADH-linked spectrophotometric assay for pyruvate dehydrogenase complex in crude tissue homogenates. J. Biol. Chem. 256, 6583–6586 (1981).
Fernandez-Vizarra, E., Lopez-Perez, M. J. & Enriquez, J. A. Isolation of biogenetically competent mitochondria from mammalian tissues and cultured cells. Methods 26, 292–297 (2002).
Acknowledgements
We thank the St Jude Cell and Tissue Imaging Facility for assistance with live-cell imaging and S. Frase for assistance with electron micrographs; the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University for Edman sequencing; B. Xia, E. Parganas and D. Gable for technical assistance; C. Shaner for animal husbandry; and members of the St Jude Biochemistry Department, S. Oakes, J. Ihle and C. Sherr for helpful discussions. J.T.O. is supported by the Pew Scholars Program in the Biomedical Sciences; the National Institutes of Health HL-102175; the American Cancer Society RSG-10-255-01-LIB; a Cancer Center Support Grant P30CA021765; and the American Lebanese Syrian Associated Charities of St Jude Children’s Research Hospital.
Author information
Authors and Affiliations
Contributions
R.M.P. and J.T.O. conceived the study, designed the experiments and wrote the manuscript. R.M.P. performed the experiments, analysed data and prepared figures. D.P.S., B.K. and M.B. generated reagents and performed experiments. J.L. and J.D.S. carried out electron-transport-chain enzymatic assays. J.T. assisted in imaging data acquisition and quantification of mitochondrial morphology. M.M.C. and R.J.Y. carried out pilot experiments on mitochondrial morphology and provided reagents. S.P. assisted in acquisition of immunofluorescence micrographs. S.M. and D.R.G. carried out experiments on oxygen consumption and provided reagents. J.T.O. supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 569 kb)
Rights and permissions
About this article
Cite this article
Perciavalle, R., Stewart, D., Koss, B. et al. Anti-apoptotic MCL-1 localizes to the mitochondrial matrix and couples mitochondrial fusion to respiration. Nat Cell Biol 14, 575–583 (2012). https://doi.org/10.1038/ncb2488
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2488
This article is cited by
-
Non-canonical BIM-regulated energy metabolism determines drug-induced liver necrosis
Cell Death & Differentiation (2024)
-
Cell-specific modulation of mitochondrial respiration and metabolism by the pro-apoptotic Bcl-2 family members Bax and Bak
Apoptosis (2024)
-
Potentiation of apoptosis in drug-resistant mantle cell lymphoma cells by MCL-1 inhibitor involves downregulation of inhibitor of apoptosis proteins
Cell Death & Disease (2023)
-
MCL1 regulates AML cells metabolism via direct interaction with HK2. Metabolic signature at onset predicts overall survival in AMLs’ patients
Leukemia (2023)
-
The mitophagy pathway and its implications in human diseases
Signal Transduction and Targeted Therapy (2023)