Abstract
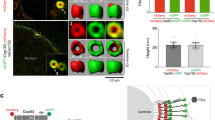
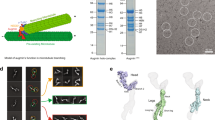
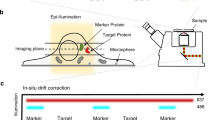
The centrosome is the main microtubule organization centre of animal cells. It is composed of a centriole pair surrounded by pericentriolar material (PCM). Traditionally described as amorphous, the architecture of the PCM is not known, although its intricate mode of assembly alludes to the presence of a functional, hierarchical structure. Here we used subdiffraction imaging to reveal organizational features of the PCM. Interphase PCM components adopt a concentric toroidal distribution of discrete diameter around centrioles. Positional mapping of multiple non-overlapping epitopes revealed that pericentrin (PCNT) is an elongated molecule extending away from the centriole. We find that PCM components occupy separable spatial domains within mitotic PCM that are maintained in the absence of microtubule nucleation complexes and further implicate PCNT and CDK5RAP2 in the organization and assembly of PCM. Globally, this work highlights the role of higher-order PCM organization in the regulation of centrosome assembly and function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Lampert, F. & Westermann, S. A blueprint for kinetochore—new insights into the molecular mechanics of cell division. Nat. Rev. Mol. Cell Biol. 12, 407–412 (2011).
Cheeseman, I. M. & Desai, A. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. 9, 33–46 (2008).
Bettencourt-Dias, M. & Glover, D. M. Centrosome biogenesis and function: centrosomics brings new understanding. Nat. Rev. 8, 451–463 (2007).
Wan, X. et al. Protein architecture of the human kinetochore microtubule attachment site. Cell 137, 672–684 (2009).
Nigg, E. A. & Raff, J. W. Centrioles, centrosomes, and cilia in health and disease. Cell 139, 663–678 (2009).
Bettencourt-Dias, M., Hildebrandt, F., Pellman, D., Woods, G. & Godinho, S.A. Centrosomes and cilia in human disease. Trends Genet. 27, 307–315 (2011).
Nigg, E. A. Centrosome aberrations: cause or consequence of cancer progression? Nat. Rev. Cancer 2, 815–825 (2002).
Kleylein-Sohn, J. et al. Plk4-induced centriole biogenesis in human cells. Dev. cell 13, 190–202 (2007).
Pelletier, L., O’Toole, E., Schwager, A., Hyman, A. A. & Muller-Reichert, T. Centriole assembly in Caenorhabditis elegans. Nature 444, 619–623 (2006).
Ou, Y. & Rattner, J. B. The centrosome in higher organisms: structure, composition, and duplication. Int. Rev. Cytol. 238, 119–182 (2004).
Dictenberg, J. B. et al. Pericentrin and γ-tubulin form a protein complex andare organized into a novel lattice at the centrosome. J. Cell Biol. 141, 163–174 (1998).
Palazzo, R. E., Vogel, J. M., Schnackenberg, B. J., Hull, D. R. & Wu, X. Centrosome maturation. Curr. Top. Dev. Biol. 49, 449–470 (2000).
Barr, F. A., Sillje, H. H. & Nigg, E. A. Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 5, 429–440 (2004).
Petronczki, M., Lenart, P. & Peters, J. M. Polo on the rise-from mitotic entry to cytokinesis with Plk1. Dev. Cell 14, 646–659 (2008).
Glover, D. M., Leibowitz, M. H., McLean, D. A. & Parry, H. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell 81, 95–105 (1995).
Glover, D. M., Hagan, I. M. & Tavares, A. A. Polo-like kinases: a team that plays throughout mitosis. Gen. Dev. 12, 3777–3787 (1998).
Hannak, E., Kirkham, M., Hyman, A. A. & Oegema, K. Aurora-A kinase is required for centrosome maturation in Caenorhabditis elegans. J. Cell Biol. 155, 1109–1116 (2001).
Moritz, M., Braunfeld, M. B., Sedat, J. W., Alberts, B. & Agard, D. A. Microtubule nucleation by γ-tubulin-containing rings in the centrosome. Nature 378, 638–640 (1995).
Zheng, Y., Wong, M. L., Alberts, B. & Mitchison, T. Nucleation of microtubule assembly by a γ-tubulin-containing ring complex. Nature 378, 578–583 (1995).
Khodjakov, A. & Rieder, C. L. The sudden recruitment of γ-tubulin to the centrosome at the onset of mitosis and its dynamic exchange throughout the cell cycle, do not require microtubules. J. Cell Biol. 146, 585–596 (1999).
Zhu, F. et al. The mammalian SPD-2 ortholog Cep192 regulates centrosome biogenesis. Curr. Biol. 18, 136–141 (2008).
Luders, J., Patel, U. K. & Stearns, T. GCP-WD is a γ-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat. Cell Biol. 8, 137–147 (2006).
Haren, L. et al. NEDD1-dependent recruitment of the γ-tubulin ring complex to the centrosome is necessary for centriole duplication and spindle assembly. J. Cell Biol. 172, 505–515 (2006).
Graser, S., Stierhof, Y. D. & Nigg, E. A. Cep68 and Cep215 (Cdk5rap2) are required for centrosome cohesion. J. Cell Sci. 120, 4321–4331 (2007).
Gomez-Ferreria, M. A. et al. Human Cep192 is required for mitotic centrosome and spindle assembly. Curr. Biol. 17, 1960–1966 (2007).
Doxsey, S. J., Stein, P., Evans, L., Calarco, P. D. & Kirschner, M. Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell 76, 639–650 (1994).
Barrera, J. A. et al. CDK5RAP2 regulates centriole engagement and cohesion in mice. Dev. Cell 18, 913–926 (2010).
Haren, L., Stearns, T. & Luders, J. Plk1-dependent recruitment of γ-tubulin complexes to mitotic centrosomes involves multiple PCM components. PLoS One 4, e5976 (2009).
Santamaria, A. et al. Use of the novel Plk1 inhibitor ZK-thiazolidinone to elucidate functions of Plk1 in early and late stages of mitosis. Mol. Biol. Cell 18, 4024–4036 (2007).
Lee, K. & Rhee, K. PLK1 phosphorylation of pericentrin initiates centrosome maturation at the onset of mitosis. J. Cell Biol. 195, 1093–1101 (2011).
Loncarek, J., Hergert, P., Magidson, V. & Khodjakov, A. Control of daughter centriole formation by the pericentriolar material. Nat. Cell Biol. 10, 322–328 (2008).
Megraw, T. L., Kao, L. R. & Kaufman, T. C. Zygotic development without functional mitotic centrosomes. Curr. Biol. 11, 116–120 (2001).
Megraw, T. L., Li, K., Kao, L. R. & Kaufman, T. C. The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development 126, 2829–2839 (1999).
Fong, K. W., Choi, Y. K., Rattner, J. B. & Qi, R. Z. CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the γ-tubulin ring complex. Mol. Biol. cell 19, 115–125 (2008).
O’Connell, K. F., Maxwell, K. N. & White, J. G. The spd-2 gene is required for polarization of the anteroposterior axis and formation of the sperm asters in the Caenorhabditis elegans zygote. Dev. Biol. 222, 55–70 (2000).
Pelletier, L. et al. The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr. Biol. 14, 863–873 (2004).
Dix, C. I. & Raff, J. W. Drosophila Spd-2 recruits PCM to the sperm centriole, but is dispensable for centriole duplication. Curr. Biol. 17, 1759–1764 (2007).
Conduit, P. T. et al. Centrioles regulate centrosome size by controlling the rate of CNN incorporation into the PCM. Curr. Biol. 20, 2178–2186 (2010).
Gopalakrishnan, J. et al. Sas-4 provides a scaffold for cytoplasmic complexes and tethers them in a centrosome. Nat. Commun. 2, 359 (2011).
Dobbie, I. M. et al. OMX: a new platform for multimodal, multichannel wide-field imaging. Cold Spring Harb. Protoc. 899–909 (2011).
Gustafsson, M. G. et al. Three-dimensional resolution doubling in wide-fieldfluorescence microscopy by structured illumination. Biophys. J. 94, 4957–4970 (2008).
Schermelleh, L. et al. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science 320, 1332–1336 (2008).
Gillingham, A. K. & Munro, S. The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 1, 524–529 (2000).
Aldaz, H., Rice, L. M., Stearns, T. & Agard, D. A. Insights into microtubulenucleation from the crystal structure of human γ-tubulin. Nature 435, 523–527 (2005).
Hutchins, J. R. et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 328, 593–599 (2010).
Hubner, N. C. et al. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J. Cell Biol. 189, 739–754 (2010).
Bolte, S. & Cordelieres, F. P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232 (2006).
Bucciarelli, E. et al. Drosophila Dgt6 interacts with Ndc80, Msps/XMAP215, and γ-tubulin to promote kinetochore-driven MT formation. Curr. Biol. 19, 1839–1845 (2009).
Goshima, G., Mayer, M., Zhang, N., Stuurman, N. & Vale, R. D. Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J. Cell Biol. 181, 421–429 (2008).
Lawo, S. et al. HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr. Biol. 19, 816–826 (2009).
Meireles, A. M., Fisher, K. H., Colombie, N., Wakefield, J. G. & Ohkura, H. Wac: a new Augmin subunit required for chromosome alignment but not for acentrosomal microtubule assembly in female meiosis. J. Cell Biol. 184, 777–784 (2009).
Uehara, R. et al. The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. Proc. Natl Acad. Sci. USA 106, 6998–7003 (2009).
Sir, J.H. et al. A primary microcephaly protein complex forms a ring around parental centrioles. Nat. Gen. 43, 1147–1153 (2011).
Wang, Z. et al. Conserved motif of CDK5RAP2 mediates its localization to centrosomes and the Golgi complex. J. Biol. Chem. 285, 22658–22665 (2010).
Zimmerman, W. C., Sillibourne, J., Rosa, J. & Doxsey, S. J. Mitosis-specific anchoring of γ tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol. Biol. Cell 15, 3642–3657 (2004).
Mahjoub, M. R., Xie, Z. & Stearns, T. Cep120 is asymmetrically localized to the daughter centriole and is essential for centriole assembly. J. Cell Biol. 191, 331–346 (2010).
Gomez-Ferreria, M. et al. Novel NEDD1 phosphorylation sites regulate γ-tubulin binding and mitotic spindle assembly. J. Cell Sci. advance online publication, http://dx.doi.org/10.1242/jcs.105130 (17 May 2012).
Joukov, V., De Nicolo, A., Rodriguez, A., Walter, J. C. & Livingston, D.M. Centrosomal protein of 192 kDa (Cep192) promotes centrosome-driven spindle assembly by engaging in organelle-specific Aurora A activation. Proc. Natl Acad. Sci. USA 107, 21022–21027 (2010).
Kim, T. et al. Novel alternatively spliced variant form of human CDK5RAP2. Cell Cycle 10, 1010–1012 (2011).
Kirkham, M., Muller-Reichert, T., Oegema, K., Grill, S. & Hyman, A.A. SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell 112, 575–587 (2003).
Bobinnec, Y. et al. Glutamylation of centriole and cytoplasmic tubulin in proliferating non-neuronal cells. Cell Motil. Cytoskeleton 39, 223–232 (1998).
Basto, R. et al. Flies without centrioles. Cell 125, 1375–1386 (2006).
Szollosi, A., Ris, H., Szollosi, D. & Debec, A. A centriole-free Drosophila cell line. A high voltage EM study. Eur. J. Cell Biol. 40, 100–104 (1986).
Mennella, V. et al. Sub-diffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat. Cell Biol. 14, http://dx.doi.org/10.1038/ncb2597 (2012).
Sonnen, K. F., Schermelleh, L., Leonhardt, H. & Nigg, E.A. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol. Open, advance online publication, http://dx.doi.org/10.1242/bio.20122337 (17 August 2012).
Kittler, R., Heninger, A. K., Franke, K., Habermann, B. & Buchholz, F. Production of endoribonuclease-prepared short interfering RNAs for gene silencing in mammalian cells. Nat. Methods 2, 779–784 (2005).
Kittler, R. et al. Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat. Cell Biol. 9, 1401–1412 (2007).
Kittler, R. et al. RNA interference rescue by bacterial artificial chromosome transgenesis in mammalian tissue culture cells. Proc. Natl Acad. Sci. USA 102, 2396–2401 (2005).
Gustafsson, M. G. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J. Microsc. 198, 82–87 (2000).
Acknowledgements
We would like to thank members of the Pelletier laboratory for stimulating discussions during the course of this work, especially C. Yeh and J. Gonçalves for their critical reading of the manuscript, C. Holley for esiRNA production, and M. Bashkurov for help with super-resolution imaging. Furthermore, we are grateful to D. Drechsel (MPI-CBG, Dresden, Germany), S. Doxsey (University of Massachusetts Medical School, Worcester, USA), M. Gomez-Ferreria (CRG, Barcelona, Spain), J. Lüders (IRB, Barcelona, Spain), K. Rhee (Seoul National University, Korea), J. Salisbury (Mayo Clinic, Minnesota, USA) and L-H. Tsai (MIT, Cambridge, USA) for providing key reagents and Applied Precision/GE Healthcare for excellent technical support with the OMX microscope. This work was financially supported by the Canadian Cancer Society (019562), the Natural Sciences and Engineering Research Council of Canada (RGPIN-355644-2008) and a grant-in-aid from the Krembil Foundation. L.P. holds a Canada Research Chair (Tier 2) in Centrosome Biogenesis and Function. S.L. is a Vanier Canada Graduate Scholar (CIHR).
Author information
Authors and Affiliations
Contributions
S.L. performed the experimental work. M.H. did interphase diameter measurements and intensity line profiles. G.D.G. performed mitotic co-localization and quantitative recruitment analysis. G.D.G. and L.P. wrote the manuscript with contributions from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 10069 kb)
Rights and permissions
About this article
Cite this article
Lawo, S., Hasegan, M., Gupta, G. et al. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat Cell Biol 14, 1148–1158 (2012). https://doi.org/10.1038/ncb2591
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2591
This article is cited by
-
Architectural basis for cylindrical self-assembly governing Plk4-mediated centriole duplication in human cells
Communications Biology (2023)
-
DNAJA2 deficiency activates cGAS-STING pathway via the induction of aberrant mitosis and chromosome instability
Nature Communications (2023)
-
An updated view on the centrosome as a cell cycle regulator
Cell Division (2022)
-
Centrosome, microtubule and DNA damage response
Genome Instability & Disease (2022)
-
Human pluripotent stem cell-derived brain organoids as in vitro models for studying neural disorders and cancer
Cell & Bioscience (2021)