Abstract
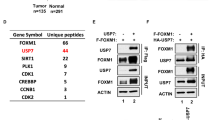
We have previously reported a gene expression signature that is a powerful predictor of poor clinical outcome in breast cancer1. Among the seventy genes in this expression profile is a gene of unknown function: TSPYL5 (TSPY-like 5, also known as KIAA1750). TSPYL5 is located within a small region at chromosome 8q22 that is frequently amplified in breast cancer, which suggests that TSPYL5 has a causal role in breast oncogenesis2,3. Here, we report that high TSPYL5 expression is an independent marker of poor outcome in breast cancer. Mass spectrometric analysis revealed that TSPYL5 interacts with ubiquitin-specific protease 7 (USP7; also known as herpesvirus-associated ubiquitin-specific protease; HAUSP). USP7 is the deubiquitylase for the p53 tumour suppressor4 and TSPYL5 reduces the activity of USP7 towards p53, resulting in increased p53 ubiquitylation. We demonstrate that TSPYL5 reduces p53 protein levels and inhibits activation of p53-target genes. Furthermore, expression of TSPYL5 overrides p53-dependent proliferation arrest and oncogene-induced senescence, and contributes to oncogenic transformation in multiple cell-based assays. Our data identify TSPYL5 as a suppressor of p53 function through its interaction with USP7.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
van 't Veer, L. J. et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 415, 530–536 (2002).
Li, Y. et al. Amplification of LAPTM4B and YWHAZ contributes to chemotherapy resistance and recurrence of breast cancer. Nat. Med. 16, 214–218 (2010).
Hu, G. et al. MTDH activation by 8q22 genomic gain promotes chemoresistance and metastasis of poor-prognosis breast cancer. Cancer Cell 15, 9–20 (2009).
Li, M. et al. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416, 648–653 (2002).
Vousden, K. H. & Lane, D. P. p53 in health and disease. Nat. Rev. Mol. Cell Biol. 8, 275–283 (2007).
Vogelstein, B., Lane, D. & Levine, A. J. Surfing the p53 network. Nature 408, 307–310 (2000).
Riley, T., Sontag, E., Chen, P. & Levine, A. Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 9, 402–412 (2008).
Brooks, C. L. & Gu, W. p53 ubiquitination: Mdm2 and beyond. Mol. Cell 21, 307–315 (2006).
van de Vijver, M. J. et al. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 347, 1999–2009 (2002).
Paik, S. et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 351, 2817–2826 (2004).
Ma, X. J. et al. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell 5, 607–616 (2004).
Sorlie, T. et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl Acad. Sci. USA 98, 10869–10874 (2001).
Fan, C. et al. Concordance among gene-expression-based predictors for breast cancer. N. Engl. J. Med. 355, 560–569 (2006).
Hu, M. et al. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell 111, 1041–1054 (2002).
Li, M., Brooks, C. L., Kon, N. & Gu, W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol. Cell 13, 879–886 (2004).
Cummins, J. M. et al. Tumour suppression: disruption of HAUSP gene stabilizes p53. Nature 428, doi:10.1038/nature02501 (2004).
Vassilev, L. T. et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303, 844–848 (2004).
Scheffner, M., Huibregtse, J. M., Vierstra, R. D. & Howley, P. M. The HPV-16 E6 and E6–AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75, 495–505 (1993).
Brummelkamp, T. R. et al. TBX-3, the gene mutated in Ulnar-Mammary Syndrome, is a negative regulator of p19ARF and inhibits senescence. J. Biol. Chem. 277, 6567–6572 (2002).
Sherr, C. J. Divorcing ARF and p53: an unsettled case. Nat. Rev. Cancer 6, 663–673 (2006).
Flatt, P. M. et al. p53-dependent expression of PIG3 during proliferation, genotoxic stress, and reversible growth arrest. Cancer Lett. 156, 63–72 (2000).
Voorhoeve, P. M. et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 124, 1169–1181 (2006).
Kuilman, T. et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell 133, 1019–1031 (2008).
Borgdorff, V. et al. Multiple microRNAs rescue from Ras-induced senescence by inhibiting p21(Waf1/Cip1). Oncogene 29, 2262–2271 (2010).
Voorhoeve, P. M. & Agami, R. The tumor-suppressive functions of the human INK4A locus. Cancer Cell 4, 311–319 (2003).
Holowaty, M. N., Sheng, Y., Nguyen, T., Arrowsmith, C. & Frappier, L. Protein interaction domains of the ubiquitin-specific protease, USP7/HAUSP. J. Biol. Chem. 278, 47753–47761 (2003).
Saridakis, V. et al. Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization. Mol. Cell 18, 25–36 (2005).
Pharoah, P. D., Day, N. E. & Caldas, C. Somatic mutations in the p53 gene and prognosis in breast cancer: a meta-analysis. Br. J. Cancer 80, 1968–1973 (1999).
Miller, L. D. et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects and patient survival. Proc. Natl Acad. Sci. USA 102, 13550–13555 (2005).
Kon, N. et al. Inactivation of HAUSP in vivo modulates p53 function. Oncogene 29, 1270–1279 (2010).
Gentleman, R. C. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80 (2004).
de Boer, E. et al. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc. Natl Acad. Sci. USA 100, 7480–7485 (2003).
Acknowledgements
We thank C. Bishop and D. Beach for the gift of HMEC cells with inducible RASV12 and members of the Bernards and Pandolfi laboratories for discussions and critical reading of the manuscript. This work was supported by grants from the Dutch Cancer Society and the Netherlands Genomics Initiative to R.B., by grants from the Netherlands Proteomics Centre and the Netherlands Genomics Initiative to J.L.B. and by NIH grants to P.P.P.
Author information
Authors and Affiliations
Contributions
The experiments were conceived and designed by M.T.E., P.P.P. and R.B. Experiments were performed by M.T.E. Mass spectrometry was performed by L.A.T.M. and supervised by J.L.B. Statistical analysis of gene expression in breast cancer was performed by O.K. The paper was written by M.T.E., P.P.P. and R.B.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 855 kb)
Rights and permissions
About this article
Cite this article
Epping, M., Meijer, L., Krijgsman, O. et al. TSPYL5 suppresses p53 levels and function by physical interaction with USP7. Nat Cell Biol 13, 102–108 (2011). https://doi.org/10.1038/ncb2142
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2142