Abstract
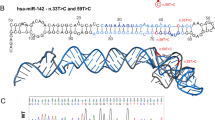
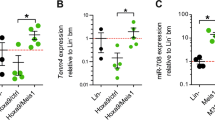
MicroRNAs (miRNAs) have emerged as novel cancer genes. In particular, the miR-17–92 cluster, containing six individual miRNAs, is highly expressed in haematopoietic cancers and promotes lymphomagenesis in vivo. Clinical use of these findings hinges on isolating the oncogenic activity within the 17–92 cluster and defining its relevant target genes. Here we show that miR-19 is sufficient to promote leukaemogenesis in Notch1-induced T-cell acute lymphoblastic leukaemia (T-ALL) in vivo. In concord with the pathogenic importance of this interaction in T-ALL, we report a novel translocation that targets the 17–92 cluster and coincides with a second rearrangement that activates Notch1. To identify the miR-19 targets responsible for its oncogenic action, we conducted a large-scale short hairpin RNA screen for genes whose knockdown can phenocopy miR-19. Strikingly, the results of this screen were enriched for miR-19 target genes, and include Bim (Bcl2L11), AMP-activated kinase (Prkaa1) and the phosphatases Pten and PP2A (Ppp2r5e). Hence, an unbiased, functional genomics approach reveals a coordinate clampdown on several regulators of phosphatidylinositol-3-OH kinase-related survival signals by the leukaemogenic miR-19.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Accession codes
References
Bartel, D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004).
Friedman, R.C., Farh, K.K., Burge, C.B. & Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105 (2009).
Grimson, A. et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27, 91–105 (2007).
Lewis, B.P., Shih, I.H., Jones-Rhoades, M.W., Bartel, D.P. & Burge, C.B. Prediction of mammalian microRNA targets. Cell 115, 787–798 (2003).
Baek, D. et al. The impact of microRNAs on protein output. Nature 455, 64–71 (2008).
Selbach, M. et al. Widespread changes in protein synthesis induced by microRNAs. Nature 455, 58–63 (2008).
Chi, S.W., Zang, J.B., Mele, A. & Darnell, R.B. Argonaute HITS-CLIP decodes microRNA–mRNA interaction maps. Nature 460, 479–486 (2009).
Xiao, C. & Rajewsky, K. MicroRNA control in the immune system: basic principles. Cell 136, 26–36 (2009).
Xiao, C. et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17–92 expression in lymphocytes. Nature Immunol. 9, 405–414 (2008).
He, L. et al. A microRNA polycistron as a potential human oncogene. Nature 435, 828–833 (2005).
Mendell, J.T. miRiad roles for the miR-17–92 cluster in development and disease. Cell 133, 217–222 (2008).
Ventura, A. et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 132, 875–886 (2008).
Mu, P. et al. Genetic dissection of the miR-17∼92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 23, 2806–2811 (2009).
Olive, V. et al. miR-19 is a key oncogenic component of mir-17–92. Genes Dev. 23, 2839–2849 (2009).
O'Donnell, K.A., Wentzel, E.A., Zeller, K.I., Dang, C.V. & Mendell, J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435, 839–843 (2005).
Petrocca, F. et al. E2F1-regulated microRNAs impair TGFβ-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell 13, 272–286 (2008).
Plas, D.R., Talapatra, S., Edinger, A.L., Rathmell, J.C. & Thompson, C.B. Akt and Bcl-xL promote growth factor-independent survival through distinct effects on mitochondrial physiology. J. Biol. Chem. 276, 12041–12048 (2001).
Malumbres, R. et al. Differentiation stage-specific expression of microRNAs in B lymphocytes and diffuse large B-cell lymphomas. Blood 113, 3754–3764 (2009).
Calin, G.A. & Croce, C.M. Investigation of microRNA alterations in leukemias and lymphomas. Methods Enzymol. 427, 193–213 (2007).
Palomero, T. et al. Activating mutations in NOTCH1 in acute myeloid leukemia and lineage switch leukemias. Leukemia 20, 1963–1966 (2006).
Ellisen, L.W. et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell 66, 649–661 (1991).
Weng, A.P. et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306, 269–271 (2004).
Pear, W.S. et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J. Exp. Med. 183, 2283–2291 (1996).
Edgar, R., Domrachev, M. & Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002).
Paddison, P.J. et al. A resource for large-scale RNA-interference-based screens in mammals. Nature 428, 427–431 (2004).
Silva, J.M. et al. Profiling essential genes in human mammary cells by multiplex RNAi screening. Science 319, 617–620 (2008).
Chang, K., Elledge, S.J. & Hannon, G.J. Lessons from Nature: microRNA-based shRNA libraries. Nature Methods 3, 707–714 (2006).
Bouillet, P. et al. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science 286, 1735–1738 (1999).
Paddison, P.J., Caudy, A.A., Sachidanandam, R. & Hannon, G.J. Short hairpin activated gene silencing in mammalian cells. Methods Mol. Biol. 265, 85–100 (2004).
John, B. et al. Human microRNA targets. PLoS Biol. 2, e363 (2004).
Mavrakis, K.J. et al. Tumorigenic activity and therapeutic inhibition of Rheb GTPase. Genes Dev. 22, 2178–2188 (2008).
Wendel, H.G. et al. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature 428, 332–337 (2004).
Yang, Y.H., Paquet, A. & Dudoit, S. R package version 1.12.0 (2006).
Tusher, V.G., Tibshirani, R. & Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA 98, 5116–5121 (2001).
Efron, B. & Tibshirani, R. On testing the significance of sets of genes. Ann. Appl. Statist. 1, 107–129 (2007).
Acknowledgements
We thank Lin He, S.W. Lowe and G. Hannon for access to shRNA and miRNA screening technologies; W. Pear, J. Huse and E.C. Holland for plasmids; H. Zhu for technical assistance; the Memorial Sloan-Kettering (MSK) animal facility and Research Animal Resource Center (RARC), A. Viale of the MSK Genomics Core, H. Zhao of the cBIO program, and J. Schatz and J. Massagué for editorial advice; and V. Murty for cytogenetic and fluorescence in situ hybridization analyses. This work was supported by grants from the American Cancer Society, the Geoffrey Beene Cancer Center, the Leukemia Research Foundation, the Louis V. Gerstner Foundation, the May and Samuel Rudin Foundation, the NY Community Trust and the William and Blanche Foundation (to H.-G.W.), from the Andrew Seligson Memorial Clinical Fellowship (to J.Z.), and NYStar (to P.J.P.). A.F. is supported by R01CA120196, the WOLF Foundation, the Rally across America Foundation and the Leukemia and Lymphoma Society (grants 1287-08 and 6237-08), and A.F. is a Leukemia & Lymphoma Society Scholar.
Author information
Authors and Affiliations
Contributions
K.J.M, A.L.W. and E.O. performed experimental design and analysis. K.d.K., T.P. and A.F. conducted T-ALL translocation analysis. K.M., J.Z., T.J. and K.C. performed the screen and analysis. A.A.K., C.S.L. and J.S.P. did data analysis. P.J.P. generated the shRNA library. W.T. was responsible for clinical specimens. H.-G.W. designed the study and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 924 kb)
Supplementary Information
Supplementary Table 1 (XLS 18 kb)
Supplementary Information
Supplementary Table 2 (XLS 31 kb)
Supplementary Information
Supplementary Table 3 (XLS 126 kb)
Supplementary Information
Supplementary Table 4 (XLS 109 kb)
Supplementary Information
Supplementary Table 5 (XLS 82 kb)
Supplementary Information
Supplementary Table 6 (XLS 1834 kb)
Supplementary Information
Supplementary Table 7 (XLS 280 kb)
Supplementary Information
Supplementary Table 8 (XLS 31 kb)
Supplementary Information
Supplementary Table 9 (XLS 30 kb)
Supplementary Information
Supplementary Table 10 (XLS 27 kb)
Supplementary Information
Supplementary Table 11 (XLS 16 kb)
Supplementary Information
Supplementary Information (PDF 105 kb)
Supplementary Information
Supplementary Information (PDF 142 kb)
Rights and permissions
About this article
Cite this article
Mavrakis, K., Wolfe, A., Oricchio, E. et al. Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia. Nat Cell Biol 12, 372–379 (2010). https://doi.org/10.1038/ncb2037
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2037
This article is cited by
-
Regulation of microRNA expression by the adaptor protein GRB2
Scientific Reports (2023)
-
BARD1 mystery: tumor suppressors are cancer susceptibility genes
BMC Cancer (2022)
-
The crucial choice of reference genes: identification of miR-191-5p for normalization of miRNAs expression in bone marrow mesenchymal stromal cell and HS27a/HS5 cell lines
Scientific Reports (2020)
-
Spermine synthase and MYC cooperate to maintain colorectal cancer cell survival by repressing Bim expression
Nature Communications (2020)
-
Regulatory network analysis reveals the oncogenesis roles of feed-forward loops and therapeutic target in T-cell acute lymphoblastic leukemia
BMC Medical Genomics (2019)