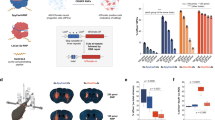
Abstract
We demonstrate editing of post-mitotic neurons in the adult mouse brain following injection of Cas9 ribonucleoprotein (RNP) complexes in the hippocampus, striatum and cortex. Engineered variants of Cas9 with multiple SV40 nuclear localization sequences enabled a tenfold increase in the efficiency of neuronal editing in vivo. These advances indicate the potential of genome editing in the brain to correct or inactivate the underlying genetic causes of neurological diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Cong, L. et al. Science 339, 819–823 (2013).
Mali, P. et al. Nat. Biotechnol. 31, 833–838 (2013).
Jinek, M. et al. Science 337, 816–821 (2012).
Jinek, M. et al. eLife 2, e00471 (2013).
Zuris, J.A. et al. Nat. Biotechnol. 33, 73–80 (2015).
Yin, H. et al. Nat. Biotechnol. 32, 551–553 (2014).
Nelson, C.E. et al. Science 351, 403–407 (2016).
Long, C. et al. Science 351, 400–403 (2016).
Tabebordbar, M. et al. Science 351, 407–411 (2016).
Wu, W.-H. et al. Mol. Ther. 24, 1388–1394 (2016).
Chew, W.L. et al. Nat. Methods 13, 868–874 (2016).
Ran, F.A. et al. Nat. Protoc. 8, 2281–2308 (2013).
Kim, S., Kim, D., Cho, S.W., Kim, J. & Kim, J.-S. Genome Res. 24, 1012–1019 (2014).
Lin, S., Staahl, B.T., Alla, R.K. & Doudna, J.A. eLife 3, e04766 (2014).
Woo, J.W. et al. Nat. Biotechnol. 33, 1162–1164 (2015).
Wang, M. et al. Proc. Natl. Acad. Sci. USA 113, 2868–2873 (2016).
Madisen, L. et al. Nat. Neurosci. 13, 133–140 (2010).
Ramakrishna, S. et al. Genome Res. 24, 1020–1027 (2014).
Dokka, S., Toledo, D., Shi, X., Castranova, V. & Rojanasakul, Y. Pharm. Res. 17, 521–525 (2000).
Armeanu, S. et al. Mol. Ther. 1, 366–375 (2000).
Liu, J., Gaj, T., Wallen, M.C. & Barbas, C.F. III. Mol. Ther. Nucleic Acids 4, e232 (2015).
Pédelacq, J.-D., Cabantous, S., Tran, T., Terwilliger, T.C. & Waldo, G.S. Nat. Biotechnol. 24, 79–88 (2006).
D'Astolfo, D.S. et al. Cell 161, 674–690 (2015).
Lewitus, G.M., Pribiag, H., Duseja, R., St-Hilaire, M. & Stellwagen, D. J. Neurosci. 34, 6146–6155 (2014).
Tiscornia, G., Singer, O. & Verma, I.M. Nat. Protoc. 1, 241–245 (2006).
Gilbert, L.A. et al. Cell 154, 442–451 (2013).
Hsu, P.D. et al. Nat. Biotechnol. 31, 827–832 (2013).
Chen, B. et al. Cell 155, 1479–1491 (2013).
Güell, M., Yang, L. & Church, G.M. Bioinformatics 30, 2968–2970 (2014).
Acknowledgements
We thank L. Harrington, T. Gaj, S. Lin, R. Rouet, C. Fellmann and D. Schaffer for productive discussions and comments on the manuscript as well as L. Bai for technical support. This work was supported by a F. Hoffmann-La Roche Postdoctoral Fellowship, RPF311, award to B.T.S. and by a Roche Pharmaceutical's Roche Alliance with Distinguished Scientists (ROADS) Fund award to J.A.D. J.A.D. is an HHMI Investigator and a Paul Allen Frontiers in Science investigator. M.B. and C.C.-B. are employed by F. Hoffmann-La Roche. A.G. was employed by F. Hoffmann-La Roche during the time of this study.
Author information
Authors and Affiliations
Contributions
B.T.S., A.G. and J.A.D. conceived the study and analyzed the data. B.T.S., J.K.S., C.U., S.N.F. and G.A.M. conducted in vitro studies. B.T.S. did Cas9 protein and sgRNA design and RNP assembly. B.T.S. and A.A.B. conducted protein production. M.B., C.C.-B. B.T.S. and J.K.S. conducted in vivo studies. B.T.S., M.B., A.G. and J.A.D. wrote the manuscript.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Corresponding author
Ethics declarations
Competing interests
The authors have submitted a patent disclosure on this work. J.A.D. is employed by HHMI and works at the University at California Berkeley. UC Berkeley and HHMI have patents pending for CRISPR technologies on which she is an inventor. J.A.D. is the executive director of the Innovative Genomics Institute at UC Berkeley and UCSF. J.A.D. is a co-founder of Editas Medicine, Intellia Therapeutics and Caribou Biosciences and a scientific advisor to Caribou, Intellia, eFFECTOR Therapeutics and Driver.
Integrated supplementary information
Supplementary Figure 1 Adaptation of Ai9/Ai14 tdTomato mouse model into a Cas9-genome editing reporter system in neural cells.
These mice harbor a modification at the Rosa26 locus with a ubiquitously expressed CAGGS promoter and a loxP-flanked stop cassette (three repeats of the SV40 polyA sequence) that prevents expression of the tdTomato fluorescent protein. This system was designed for Cre-mediated recombination at the loxP sites leading to excision of the stop cassette and tdTomato expression. This mouse model provides a robust, high-throughput, quantitative readout of site-specific genome modification at the loxP-flanked stop cassette locus with a gain-of-function fluorescent signal in modified cells17. However, one of the two loxP sites lacks a Protospacer Adjacent Motif (PAM) site necessary for S. pyogenes Cas9-mediated DNA cleavage, and therefore, two unique single-guide RNAs (sgRNAs) would be needed for Cas9 to cut as Cre does at the loxP sites. Therefore we set out to find a unique targetable sequence within the stop cassette that is capable of activating tdTomato expression. To test this we isolated E13.5 Neural Progenitor/Stem cells (NPCs) from homozygous tdTomato mice. We nucleofected these cultures with plasmids encoding S. pyogenes Cas9 and various sgRNA’s that targeted the stop cassette. From this experiment we determined a stop cassette-targeting sgRNA, sgRNA298 hereafter referred to as sgRNAtdTom, which most efficiently activated tdTomato expression. A) Experimental scheme of Cas9 RNP delivery in vitro or in vivo for genome editing of tdTomato locus and genetic or phenotypic characterization. Coronal brain sections with native tdTomato signal in edited cells. Scale bar 1mm. B) Location of sgRNA’s on tdTomato stop cassette. C) Cas9;sgRNA target sites differentially activate tdTomato. sgRNAs organized on histogram by 5’-3’ position on tdTomato stop cassette. D) Table of sgRNA sequences with Off target hit scores26. E) Neural Progenitor Cell cultures are Nestin+, a neural progenitor/stem cell marker protein. Scale bar = 100μm.
Supplementary Figure 2 Cas9 RNP-mediated editing of Ai9 mouse tdTomato stop cassette.
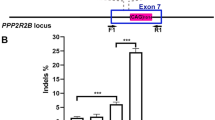
A) Cartoon illustrating tdTomato stop cassette locus before and after Cas9 editing. sgRNA-tdTom (aka target298) has three target sites in the stop cassette (red boxes) and therefore can generate six types of edits, including three different small 1-2bp indels that do not generate a deletion, two types of single-repeat deletions and one double-repeat deletion. The tdTomato NPCs are homozygous with two tdTomato reporter genes per cell. The double repeat deletion activated tdTomato expression. PCR primers used for genomicDNA PCR, 272F/273R. B) Quantification of activation of native tdTomato protein by flow cytometry and editing efficiency by NexGen Sequencing28 in NPCs, primers 344F/345R. n=3 independent experiment replicates for flow cytometry analysis. C) PCR analysis of edited stop cassette, 3 days post RNP nucleofection reveals the expected triple DNA laddering pattern in edited cells; Top band = unedited or small 1-2bp indels, middle band = single repeat deletion, bottom band = double repeat deletion pattern. These three observed bands correspond to the 6 expected products due to the shared sizes of some of the products. To determine which type of edits activate tdTomato expression we used fluorescence-activated cell sorting (FACS) to separate fluorescent tdTomato+ (RFP+) and non-fluorescent (RFP-) cells within samples of 10pmol RNP nucleofected NPCs in which 39% +/-2% of the cells are tdTomato+ (Supplementary Fig. 2B). PCR analysis of FACS sorted RFP- and RFP+ cells, indicates that only double repeat deletion/bottom band in stop cassette activates tdTomato RFP, whereas the single repeat deletion occurred in both RFP+ and RFP- cell populations. Therefore, activation of tdTomato required double deletion that removed two of three SV40 polyA repeat sequences, in at least one of two reporter alleles, while single repeat deletions did not activate tdTomato. D) Sanger sequencing of 100 clones of the top DNA band from RFP- FACS sorted cells revealed that 45% of these alleles contained small 1-2 bp indels at 1, 2 or 3 of the sgRNA298 target sites that did not activate tdTomato expression. E) Representative WT and edited alleles amplified from genomicDNA with primers 272F/273R. DNA bands were gel purified, cloned, Sanger sequenced and aligned to tdTomato STOP cassette. Based on these data, we estimate that 80% of alleles ([0.45 x 36%] + 28% + 36% = 80.2%) in the 10pmol RNP nucleofected samples were edited, with ~34+/-2% of the alleles acquiring the multiple edits necessary to activate tdTomato expression. This correlates well with 39% +/-2% tdTomato+ cells in 10pmol RNP nucleofected samples (Supplementary Fig. 2B). The correlation of tdTomato RFP reporter activation and genomicDNA quantitative PCR-based analysis of the bottom DNA band, validates the quantitative PCR analysis.
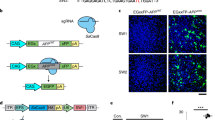
Supplementary Figure 3 Cas9 RNP variants are equally active in mitotic and post-mitotic cells when assisted across the cell membrane but only 4xNLS-Cas9-2xNLS is active in post-mitotic cortical neurons under direct delivery conditions.
A) Cas9 RNP complexes targeting d2EGFP are mixed with Lipofectamine2000 and delivered to HEK293T-d2EGFP cells. Percent EGFP gene disruption is measured by FACS analysis. RNPs mixed with Lipofectamine2000 resulted in robust GFP disruption and 0xNLS-Cas9-2xNLS was not significantly different from 4xNLS-Cas9-2xNLS at all RNP doses. Data are represented as mean ± SD (4xNLS-Cas9;sgRNA-NT3 vs. 0xNLS-Cas9;sgRNA-NT3; Two-tailed Unpaired t test with equal SD; p = 0.9032 and F5, 5 = 1.034). n=3 experimental replicates with 3 technical replicates each. B) NaCl hypertonic protein transduction protocol to deliver RNPs into tdTomato NPCs23. Editing activity for 4xNLS-Cas9-2xNLS-sfGFP, 0xNLS-Cas9-2xNLS-sfGFP, 0xNLS-Cas9-2xNLS and Cas9 with no NLS was not significantly different at all RNP doses. (2way ANOVA with Tukey’s multiple comparisons test; p=0.8685 and F9,19=0.4825). n=3 experimental replicates with 2 technical replicates each. C) We assessed the ability of 4xNLS-Cas9-2xNLS and 0xNLS-Cas9-2xNLS to edit primary cortical neurons (which are post-mitotic and no longer breakdown the nuclear envelope during cell division) in cell culture by direct delivery of RNP complexes or by LNP carrier assisted delivery, RNP+Lipofectamine2000. The efficiency of neuron editing was low in all cases, <0.05% of all seeded cells were tdTomato+ / MAP2+ or NeuN+ (marker proteins of post-mitotic mature neurons). Direct delivery of 4xNLS-Cas9-2xNLS results in edited post-mitotic neurons (NeuN+/tdTomato+) while 0x-NLS-Cas9-2xNLS does not. n=3 experimental replicates with 2 technical replicates each. D) In contrast, when RNPs were mixed with Lipofectamine2000 both 4xNLS-Cas9-2xNLS and 0xNLS-Cas9-2xNLS RNPs triggered genome editing in mature neurons (MAP2+, tdTomato+). Therefore, both 4xNLS-Cas9-2xNLS and 0xNLS-Cas9-2xNLS RNPs are capable of editing post-mitotic neurons if assisted across the cell membrane by Lipofectamine2000, but only 4xNLS-Cas9-2xNLS is capable of penetrating and triggering genome editing in neurons when directly added to the media. n=3 experimental replicates with 2 technical replicates each. Scale bar 10μm.
Supplementary Figure 4 Bilateral intrastriatal injection measurements of tdTomato+ cell volume and density indicates RNP dose dependent increase in edited tissue volume.
A) Red oval indicates region of tdTomato+ cells on sagittal and coronal cartoons of the mouse brain. Injection needle with 200μm outer diameter is drawn for reference. Dashed lines on sagittal section represent approximate positions of 50μm coronal sections along the rostral-caudal axis. B) Representative images of serial coronal sections from a 30pmol 4xNLS-Cas9-2xNLS injected striatum are presented here with approximate Bregma coordinates. With serial sectioning at the periodicity of 1 in 6, each coronal section represented here samples 300μm of tissue making the volume of edited cells 1.5mm3. Asterisks (*) indicate position of tissue sections used for Laser microdissection of tdTomato+ dorsal striatal tissue. Scale bar = 1.4mm. C) Area representation with # of tdTomato+ cells on Y-axis and rostral-caudal position of coronal sections analyzed on X-axis. X-axis units are millimeters (mm). 4pmol 0xNLS-Cas9-2xNLS RNP edits fewer cells and extends for ~1mm along rostral-caudal axis. Density and volume of tdTomato+ cells along rostral-caudal axis increases with increasing dose of 4xNLS-Cas9-2xNLS. n=3 animals, n=2 bilateral injections per animal for each group. Data are represented as mean ± SEM.
Supplementary Figure 5 Quantification of in vivo editing efficiency by genomicDNA PCR analysis of tdTomato stop locus.
A) Laser microdissection of tissue from 30pmol RNP treated dorsal striatum was used for this analysis. Rectangles of tissue containing tdTomato+ cells, ~1mm x ~1.5mm, were microdissected from three 50μm thick sections spanning ~1mm along rostral-caudal axis (marked with asterisk in Supplementary Fig. 4B) and combined. PCR analysis of genomic DNA isolated from this tissue confirmed the expected deletions in 7.18% +/-2.18% of the alleles of which 3.7%+/-0.18% of alleles have the double deletion edit that activates tdTomato expression. n=3 animals, n=2 bilateral injections per animal for each group. B) Sanger sequencing of the top band PCR product (corresponding in size to wild-type and/or small 1-2bp indel editing events) revealed an additional 8.8% of alleles with small 1-2bp indels at 1,2, or 3 target sites, as was observed for tdTomato mouse NPCs edited in vitro. Based on this analysis we estimate tdTomato+ cells is reporting ~23% of the total editing in the dorsal striatum. (tdTomato+ “double deletion” alleles / total edited alleles i.e. 3.7% / (7.18% (deletion edits) + 8.8% (small indels)=16%) = 23%. Therefore we can extrapolate that 30pmol 4xNLS-Cas9-2xNLS RNP edits ~4x as many alleles as tdTomato+ cells report, i.e. 2675 +/-613 tdTomato+ cells / 0.23 = 11,630 cells in ~1.5mm3 volume of tissue.
Supplementary Figure 6 Analysis of innate immune response in sham treated, RNP treated and untreated brains.
A) Morphological appearance of microglia in the mouse striatum in Cas9 RNP complex RNP treated and untreated mice visualized in green by immunostaining with anti-Iba-1 antibody. Iba-1 (ionized calcium-binding adapter molecule 1, also known as Allograft inflammatory factor 1 (AIF-1)) is useful as an indicator of activated microglia because 1) its levels increase and 2) the cytoplasmic staining pattern can be used to assess microglia morphological changes that occur upon activation, i.e. microglia enlarge their cell bodies and thicken their processes, which closely enwrap neuronal cell bodies29. In Cas9 RNP treated mice, microglia have small cell bodies and long and slender processes indicating they are not activated. B) Quantification of Iba-1 protein intensity in untreated, sham (buffer only) and 4x-NLS-Cas9-2xNLS treated mice. Data are represented as mean ± SEM with scatter plots of mean intensity of 2-8 sections per animal (One way ANOVA; p = 0.4496 and F2, 7 = 0.898). n=3 animals per condition with 2-8 sections analyzed per animal. For each section, fluorescence intensity of the Iba-1 labeled channel for the entire 20X field was measured with image J. The experimenter was blinded to treatment condition while performing quantitation of fluorescence intensity. C) A panel of microglia markers of activation were analyzed at two timepoints, 3 and 12 days post injection by qPCR transcript analysis. The panel contains: CD11b, CD45, CD68, CD86, Cx3cr1, IBA-1, IL-12b p40, IL-12a p35, Tmem119 and TNF-α. Dorsal striatum of sham injected (buffer only) and Cas9 RNP (50pmol/0.5ul) injected mice were collected at 3 days post injection (n=3 animals, n=2 bilateral injections per animal) and 12 days post injection (n=3 animals, n=2 bilateral injections per animal). Transcripts that are significantly different in sham vs. RNP injected at 3 days post injection are Cd11b, Cx3cr1, IL-12b p40 and IL-12a p35. All are depleted in RNP-treated samples relative to sham-treated. At 12-day post injection, TNF-α is the only transcript significantly different and increased 5.65-fold in RNP-treated relative to sham-treated samples. Data are presented as fold change relative to sham for respective timepoint. Data are represented as mean ± SEM with scatter plots of individual samples. Data for 3 days post injection. Two-tailed unpaired t test with equal SD was used to assess significance for the following genes: Cd11b p=0.0378 and F5,5 2.903, CD45 p=0.1298 and F5,5 2.133, CD68 p=0.0718 and F5,5 32.36, CD86 p=0.3150 and F5,5 1.22, Cx3cr1 p=0.0371 and F5,5 12239, IBA-1 p=0.1100 and F5,5 6.714, IL-12b p40 p=0.0322 and F5,5 3.71, IL-12a p35 p=0.0138 and F5,5 4.44, Tmem119 p=0.1946 and F5,5 1.518, TNF-α p=0.9248 and F5,5 2.776. Data for 12 days post injection. Two-tailed unpaired t test with equal SD was used to assess significance for the following genes: Cd11b p=0.1072 and F5,5 7.168, CD45 p=0.4906 and F5,5 2.177, CD68 p=0.0562 and F5,5 10.72, Cx3cr1 p=0.1548 and F5,5 217, IBA-1 p=0.3129 and F5,5 3.355, IL-12b p40 p=0.0625 and F5,5 4.272, IL-12a p35 p=0.0657 and F5,5 14.54, P2ry12 p=0.0760 and F5,5 67.41, TNF-α p=0.0384 and F5,5 23.81.
Supplementary information
Supplementary Text and Figures
Supplementary figures 1–6 (PDF 1325 kb)
Rights and permissions
About this article
Cite this article
Staahl, B., Benekareddy, M., Coulon-Bainier, C. et al. Efficient genome editing in the mouse brain by local delivery of engineered Cas9 ribonucleoprotein complexes. Nat Biotechnol 35, 431–434 (2017). https://doi.org/10.1038/nbt.3806
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.3806
This article is cited by
-
Acoustically targeted noninvasive gene therapy in large brain volumes
Gene Therapy (2024)
-
Efficient engineering of human and mouse primary cells using peptide-assisted genome editing
Nature Biotechnology (2024)
-
Engineering self-deliverable ribonucleoproteins for genome editing in the brain
Nature Communications (2024)
-
Reprogramming an RNA-guided archaeal TnpB endonuclease for genome editing
Cell Discovery (2023)
-
Combinatorial design of nanoparticles for pulmonary mRNA delivery and genome editing
Nature Biotechnology (2023)