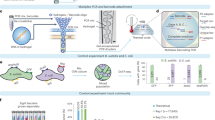
Abstract
Methods to rapidly assess cell growth would be useful for many applications, including drug susceptibility testing, but current technologies have limited sensitivity or throughput. Here we present an approach to precisely and rapidly measure growth rates of many individual cells simultaneously. We flow cells in suspension through a microfluidic channel with 10–12 resonant mass sensors distributed along its length, weighing each cell repeatedly over the 4–20 min it spends in the channel. Because multiple cells traverse the channel at the same time, we obtain growth rates for >60 cells/h with a resolution of 0.2 pg/h for mammalian cells and 0.02 pg/h for bacteria. We measure the growth of single lymphocytic cells, mouse and human T cells, primary human leukemia cells, yeast, Escherichia coli and Enterococcus faecalis. Our system reveals subpopulations of cells with divergent growth kinetics and enables assessment of cellular responses to antibiotics and antimicrobial peptides within minutes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Labhsetwar, P., Cole, J.A., Roberts, E., Price, N.D. & Luthey-Schulten, Z.A. Heterogeneity in protein expression induces metabolic variability in a modeled Escherichia coli population. Proc. Natl. Acad. Sci. USA 110, 14006–14011 (2013).
Balaban, N.Q., Merrin, J., Chait, R., Kowalik, L. & Leibler, S. Bacterial persistence as a phenotypic switch. Science 305, 1622–1625 (2004).
Di Talia, S., Skotheim, J.M., Bean, J.M., Siggia, E.D. & Cross, F.R. The effects of molecular noise and size control on variability in the budding yeast cell cycle. Nature 448, 947–951 (2007).
van Heerden, J.H. et al. Lost in transition: start-up of glycolysis yields subpopulations of nongrowing cells. Science 343, 1245114 (2014).
Sandler, O. et al. Lineage correlations of single cell division time as a probe of cell-cycle dynamics. Nature 519, 468–471 (2015).
Reya, T., Morrison, S.J., Clarke, M.F. & Weissman, I.L. Stem cells, cancer, and cancer stem cells. Nature 414, 105–111 (2001).
Keren, I., Kaldalu, N., Spoering, A., Wang, Y. & Lewis, K. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230, 13–18 (2004).
Fridman, O., Goldberg, A., Ronin, I., Shoresh, N. & Balaban, N.Q. Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature 513, 418–421 (2014).
Aldridge, B.B. et al. Asymmetry and aging of mycobacterial cells lead to variable growth and antibiotic susceptibility. Science 335, 100–104 (2012).
Wang, P. et al. Robust growth of Escherichia coli. Curr. Biol. 20, 1099–1103 (2010).
Moffitt, J.R., Lee, J.B. & Cluzel, P. The single-cell chemostat: an agarose-based, microfluidic device for high-throughput, single-cell studies of bacteria and bacterial communities. Lab Chip 12, 1487–1494 (2012).
Taheri-Araghi, S. et al. Cell-size control and homeostasis in bacteria. Curr. Biol. 25, 385–391 (2015).
Ferrezuelo, F. et al. The critical size is set at a single-cell level by growth rate to attain homeostasis and adaptation. Nat. Commun. 3, 1012 (2012).
Barer, R. Interference microscopy and mass determination. Nature 169, 366–367 (1952).
Zangle, T.A. & Teitell, M.A. Live-cell mass profiling: an emerging approach in quantitative biophysics. Nat. Methods 11, 1221–1228 (2014).
Godin, M. et al. Using buoyant mass to measure the growth of single cells. Nat. Methods 7, 387–390 (2010).
Son, S. et al. Direct observation of mammalian cell growth and size regulation. Nat. Methods 9, 910–912 (2012).
Park, K. et al. Measurement of adherent cell mass and growth. Proc. Natl. Acad. Sci. USA 107, 20691–20696 (2010).
Burg, T.P. et al. Vacuum-packaged suspended microchannel resonant mass sensor for biomolecular detection. J. Microelectromech. Syst. 15, 1466–1476 (2006).
Feijó Delgado, F. et al. Intracellular water exchange for measuring the dry mass, water mass and changes in chemical composition of living cells. PLoS One 8, e67590 (2013).
Lee, J., Bryan, A.K. & Manalis, S.R. High precision particle mass sensing using microchannel resonators in the second vibration mode. Rev. Sci. Instrum. 82, 023704 (2011).
Lee, J. et al. Suspended microchannel resonators with piezoresistive sensors. Lab Chip 11, 645–651 (2011).
Lee, J., Shen, W., Payer, K., Burg, T.P. & Manalis, S.R. Toward attogram mass measurements in solution with suspended nanochannel resonators. Nano Lett. 10, 2537–2542 (2010).
Carson, J.R. Notes on the theory of modulation. Proc. Inst. Radio Eng. 10, 57–64 (1922).
Tortonese, M., Yamada, H., Barrett, R.C. & Quate, C.F. Atomic force microscopy using a piezoresistive cantilever. Transducers 1991, 448–451 (1991).
Olcum, S., Cermak, N., Wasserman, S.C. & Manalis, S.R. High-speed multiple-mode mass-sensing resolves dynamic nanoscale mass distributions. Nat. Commun. 6, 7070 (2015).
Kuhn, H.W. The Hungarian method for the assignment problem. Nav. Res. Logist. Q. 2, 83–97 (1955).
Allan, D.W. Statistics of atomic frequency standards. Proc. IEEE 54, 221–230 (1966).
Fox, C.J., Hammerman, P.S. & Thompson, C.B. Fuel feeds function: 934energy metabolism and the T-cell response. Nat. Rev. Immunol. 5, 844–852 (2005).
Halpern, B. & Amache, N. Diagnosis of drug allergy in vitro with the lymphocyte transformation test. J. Allergy 40, 168–181 (1967).
Poujol, F. et al. Flow cytometric evaluation of lymphocyte transformation test based on 5-ethynyl-2′deoxyuridine incorporation as a clinical alternative to tritiated thymidine uptake measurement. J. Immunol. Methods 415, 71–79 (2014).
Elkord, E., Williams, P.E., Kynaston, H. & Rowbottom, A.W. Human monocyte isolation methods influence cytokine production from in vitro–generated dendritic cells. Immunology 114, 204–212 (2005).
Andreu, D. et al. Shortened cecropin A-melittin hybrids. Significant size reduction retains potent antibiotic activity. FEBS Lett. 296, 190–194 (1992).
Fantner, G.E., Barbero, R.J., Gray, D.S. & Belcher, A.M. Kinetics of antimicrobial peptide activity measured on individual bacterial cells using high-speed atomic force microscopy. Nat. Nanotechnol. 5, 280–285 (2010).
Mir, M. et al. Optical measurement of cycle-dependent cell growth. Proc. Natl. Acad. Sci. USA 108, 13124–13129 (2011).
Burg, T.P. et al. Weighing of biomolecules, single cells and single nanoparticles in fluid. Nature 446, 1066–1069 (2007).
Olcum, S. et al. Weighing nanoparticles in solution at the attogram scale. Proc. Natl. Acad. Sci. USA 111, 1310–1315 (2014).
Stone, H.A. in CMOS Biotechnology (eds. Lee, H., Westervelt, R.M. & Ham, D.) 5–30 (Springer US, 2007).
Savitzky, A. & Golay, M.J.E. Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 36, 1627–1639 (1964).
Lane, A.A. et al. Triplication of a 21q22 region contributes to B cell transformation through HMGN1 overexpression and loss of histone H3 Lys27 trimethylation. Nat. Genet. 46, 618–623 (2014).
Acknowledgements
We thank G. Szeto for providing mouse spleen and lymph nodes, J.H. Kang for measuring the density of RPMI 1640 and A. Amon for a critical reading of the manuscript. This work was carried out in part through the use of the Microsystems Technology Laboratories at the Massachusetts Institute of Technology (MIT). Support was provided by the Bridge Project (a partnership between the Koch Institute for Integrative Cancer Research at MIT and the Dana-Farber/Harvard Cancer Center) to S.R.M. and D.M.W.; the US Army Research Office (Institute for Collaborative Biotechnologies grant W911NF-09-D-0001) to S.R.M.; the US National Science Foundation (1129359) to S.R.M.; Institut Mérieux to S.R.M.; and the US National Cancer Institute (Physical Sciences Oncology Center U54CA143874 and Cancer Center Support grants P30-CA14051 and R33-CA191143) to S.R.M. N.C. acknowledges support from an MIT Poitras Fellowship. M.A.M. acknowledges support from NCI institutional research training grant T32 CA009172, awarded to the Department of Medical Oncology at Dana-Farber Cancer Institute. M.M.S. acknowledges support from the NIH/NIGMS T32 GM008334 Interdepartmental Biotechnology Training Program grant. D.M.W. is a Leukemia and Lymphoma Society Scholar.
Author information
Authors and Affiliations
Contributions
N.C., S.O. and F.F.D. designed devices; N.C., S.O. and S.C.W. designed and constructed the experimental setup; K.R.P., M.O., V.A. and F.B. fabricated the devices; N.C., S.O., M.M.S., R.J.K., S.M.K., A.S., Y.K., D.M.W. and S.R.M. designed the experiments, N.C., S.O., S.M.K., R.J.K., M.A.M. and M.M.S. performed the experiments; N.C. analyzed the data; N.C., S.O. and S.R.M. wrote the paper with input from all authors.
Corresponding author
Ethics declarations
Competing interests
S.R.M. is a cofounder of Affinity Biosensors, which develops techniques relevant to the research presented. S.O. and M.M.S. anticipate employment at Affinity Biosensors. The Massachusetts Institute of Technology has filed a patent (USPTO 9,134,295) covering part of the information contained in this article.
Integrated supplementary information
Supplementary Figure 1 Schematic of the control system for the serial microfluidic mass sensor array.
(A) Schematic of optical and electronic path of parallel feedback loops for each mass sensor. (B) Photograph of the optical setup implementing the schematic in (A). The photograph shows the sample holder and the fluidic connections to the sensor chip.
Supplementary Figure 2 Transfer functions of the PLL–cantilever control loops.
Measured transfer functions (colored lines) of all twelve PLL-cantilever feedback loops on a single large-channel serial SMR device. Bandwidth had been set to 100 Hz according to the method in [1]. Solid black line shows ideal 100 Hz first-order response, dashed grey line indicates -3 dB bandwidth.
Supplementary Figure 3 Sensitivities of the mass sensors in a large-channel device.
Resonant frequency versus mass sensitivity for a single large-channel device, measured daily during the mouse CD8 cell experiments (Figure 3 in the main text). Dashed grey line shows best fit of data to y = ax1.5, illustrating how sensitivity scales with frequency to the power of 1.5. While fabrication tolerances and slight variations in geometry may explain some of the deviations from the model, it remains unclear why some cantilevers exhibit substantial day-to-day variation.
Supplementary Figure 4 Example contour plots of the cost function used in the matching algorithm.
Contour plots of the cost function for several simulated example cells with varying numbers of previous peaks observed (black points). (A) If a cell only has a single peak assigned, the cost function is shaped like a wide bowl, shaped almost entirely by the prior assumptions on mass accumulation rate and system noise. (B), (C) As more data points are observed, the mass accumulation rate becomes established and the cost function contours become determined by the system’s mass measurement noise.
Supplementary Figure 5 Simulation of the cell-matching process.
Simulation of the cell matching process showing that single cells are reliably matched by our method. (A) We simulate a set of cells sampled from a joint distribution of mass and mass accumulation rate similar to the L1210 cells shown in Figure 2. However, we simulate cells entering the serial SMR array at a rate of 100 cells per hour (two-fold more concentrated than we have used in our experiments). Each cell varies in the time it takes to travel from each cantilever to the next (mean 1.9 minutes, standard deviation 0.3 minutes), and Gaussian noise is added to each buoyant mass measurement (standard deviation 0.05 pg, similar to that of our large-channel device). (B) We then match the measurements in the simulated data. All data points that have been matched together as corresponding to the same cell have been colored the same randomly-chosen color. (C) Comparison of the masses and mass accumulation rates from which the data in (A) was generated, and the observed mass and mass accumulation rates, showing excellent agreement. (D) Comparison of mass accumulation rates from which the data in (A) was generated, and the observed mass accumulation rates, showing excellent agreement, except for in the case of two mismatched cells (off-diagonal points) out of 300 in the simulated dataset.
Supplementary Figure 6 Stability of the cantilevers used in the mass sensor arrays.
Measured Allan deviations of all cantilevers on two separate devices. Left two plots show Allan deviations in fractional frequency units, relative to the unloaded cantilever frequency (colored dots/lines). For reference, the dashed grey line indicates the measured noise performance of an optimized piezoresistive single large-channel SMR. Right two plots show Allan deviations rescaled by each cantilever’s mass sensitivity. Theoretical thermomechanical limitations on the lowest achievable Allan deviations are also plotted (calculated from [2]), assuming the cantilever is driven to a mean-squared displacement one billion times (90 dB) above the thermally-driven mean-squared displacement. While larger drive amplitudes would theoretically further reduce these limits, mechanical nonlinearity typically becomes significant beyond 90 dB, limiting noise performance.
Supplementary Figure 7 Mass accumulation rate resolution of the large-channel devices.
Measuring a mixture of plastic microparticles to determine mass accumulation rate resolution on a large-channel serial mass sensor array. (A) We measured a mixture of 4, 6, 7, 8, 9, 10 and 12 μm polystyrene beads (Duke Standards, NIST traceable, Thermo Scientific) at 37 C in 0.01% Tween-20 in water. Sensors were calibrated by linearly rescaling their raw frequency signals such that the 7 μm bead modal mass is the expected buoyant mass (10.15 pg). (B) Across all sizes and sensors, particle buoyant masses match the expected buoyant masses (dashed lines), verifying that the sensors are linear over this size range. (C) 4 μm particles have the lowest size variability (in pg) of these beads according to manufacturer’s datasheet, and therefore their distribution’s width is a reasonable upper bound on the sensor error. Typical sensor root-mean-square-error is on the order of 0.05 pg. (D) Histogram of mass accumulation rates (errors, as particles are not growing) of 85 single particles for which at least 10 sensors could be linked together. Mass accumulation rates were calculated excluding data from the first sensor, which displayed much higher noise than the other sensors. Dashed line shows estimated mass accumulation rate distribution assuming t = 1.4 minutes, k = 11, and s = 0.05 pg, showing good agreement between this approximation for mass accumulation rate error and the observed mass accumulation rate error distribution.
Supplementary Figure 8 Mass accumulation rate resolution of the small-channel devices.
Same as Supplementary Figure 7, but for a small-channel serial SMR array. (A) We measured a mixture of 1.0, 1.36, 1.57, 1.74, 2.0, and 2.5 μm polystyrene beads (Duke Standards, Thermo Scientific, except for 1.0 μm, from Bangs Labs) at 37 C in LB with 0.1% Tween-80. Sensors were calibrated by linearly rescaling their raw frequency signals such that the 1.0 μm bead modal buoyant mass is 0.02 pg (the expected buoyant mass in LB at a density of 1.013 g/mL). (B) Distributions of measured buoyant masses for each SMR in the array, demonstrating both linearity and precision in each cantilever mass measurement. (C) Buoyant mass distributions of 1 μm polystyrene particles provide an upper bound on each cantilever’s buoyant mass measurement error, here on the order of 0.5-1 fg. (D) Mass accumulation rate distribution of single 362 single particles, yielding a mass accumulation rate standard error of 0.022 pg/h. Dashed line shows mass accumulation rate distribution based on equation 2 assuming t = 24 seconds, k = 10, and σ = 0.001 pg.
Supplementary Figure 9 Mass accumulation rate of a fixed mouse lymphoblast cell line (L1210).
(A) Buoyant mass trajectories for fixed L1210 cells, measured in phosphate-buffered saline at 37 C. (B) By plotting mass accumulation rate against time, the first cells going through the array can be seen to be losing mass. We believe this is real mass loss attributable to the temperature shift (cells had been fixed and stored at 4 C), and note that after one hour into the measurement cells appear to have equilibrated and no further mass loss occurs. (C) Histogram of mass accumulation rates of fixed cells, excluding the first hour of measurements.
Supplementary Figure 10 FACS plots of primary acute myeloid leukemia cells.
FACS plots of two primary AML samples whose growth properties were assessed on the SMR. Fresh primary peripheral blood or bone marrow samples from patients with newly diagnosed AML were subject to erythrocyte lysis and stained with antibodies targeting human CD33 and human CD15, and leukemia cells were enriched by performing FACS for hCD33/hCD15 double-positive cells. Left panel (sample 1): primary peripheral blood sample from a patient with AML with monocytic differentiation and extensive circulating disease. Contemporaneous clinical testing confirmed that this leukemia expressed CD33 and CD15 and demonstrated that it comprised 43% of peripheral blood mononuclear cells, on which this sample was gated. Of note, this specimen was obtained after the patient had received cytoreductive chemotherapy (hydroxyurea) for three days. Right panel (sample 2): primary bone marrow aspirate from a patient with therapy-related AML. Of note, this specimen did not undergo immunophenotyping by the clinical lab, but morphologic analysis suggested that the leukemia comprised a minority of cells in this double-positive population.
Supplementary Figure 11 Culturing AML cells ex vivo.
We seeded cells obtained from Patient 1 (Figure 4 and Supplementary Figure 10) at 0.5, 0.7 and 1.5 million cells/mL (blue, red, and black, respectively) into 6-well culture dishes. (A) We profiled these cultures volume distributions over the next 48 hours with a coulter counter (Beckman Coulter Multisizer 4, 100 μm aperture). The cultures behaved similarly for different inoculation densities. (B) Total cell counts generally increased slightly, but total biovolume decreased.
Supplementary Figure 12 Mass accumulation rate of E. faecalis measured on a small-channel mass sensor array.
At the left, colored dots show points which were determined to correspond to a single cell, for which the mass accumulation rate was determined and plotted against the cell’s mass (right). Grey points indicate measurements for which less than seven mass measurements could be linked together, and were not used in the analysis at right. E. faecalis was grown in Brain-Heart Infusion (BHI, Difco) overnight and transferred to fresh BHI with 0.2% Tween-80 at a 105-fold dilution approximately three hours prior to measurement. 1.36 μm beads were used as the calibration standard and have been omitted from the plot at left for clarity.
Supplementary Figure 13 Comparison of precision between recent quantitative-phase microscopy measurements and SMR measurements.
Plots in (A) and (B) are excerpted from Mir et al. [3]. Insets in the original figure (A) and caption segment describing the insets have been omitted for clarity. Original caption reads:
SLIM measurements of E. coli growth. (A) Dry mass vs. time for a cell family. [...] The blue line is a fixed cell measurement, with SD of 19.6 fg. Markers indicate raw data, and solid lines indicate averaged data. (B) Growth rate vs. mass of 20 cells measured in the same manner. Faint circles indicate single data points from individual cell growth curves, dark squares show the average, and the dashed line is a linear fit through the averaged data; the slope of this line, 0.011 min 1, is a measure of the average growth constant for this population. The linear relationship between the growth rate and mass indicates that, on average, E. coli cells exhibit exponential growth behavior.
(C) Single-cell E. coli (ATCC 43893) growth trajectories measured on a single SMR (160 μm long with a 3 by 5 μm interior channel, operated in the second vibrational mode at 1.1 MHz) of similar design to the SMRs in serial SMR arrays. Growth was measured by passing a single cell back and forth through the SMR, as in [4]. Data points from other cells that entered the sensor during the dynamic trap (but were ignored by the trapping algorithm) were removed. SMR measurements were made in LB at room temperature, yielding a similar growth rate as in Mir et al. [3], which used E. coli MG1655 in M9-casamino acid media at 37 C. (D) Colored points are buoyant mass accumulation rates estimated from the data in (C), based on linear fits to non-overlapping 5-minute segments. Five minutes was chosen as that was the width of the applied smoothing filter in (A) and (B). Black points are mass accumulation rates from E. coli cells at 37 C measured in the serial SMR array in Figure 6A. Dashed lines show best linear fits in which the intercept was forced to zero, and corresponding exponential growth rates are noted for the two experiments. Note that the terminology of ’growth rate’ used in (B) is equivalent to ’mass accumulation rate’.
Supplementary Figure 14 Theoretical trade-off between throughput and resolution.
Theoretical trade-off between throughput and resolution for the large-channel devices used in this study, with 12 mass sensors and delay channels with volumes 120-fold higher than the volume of a single cantilever. We assume the cell concentration cannot exceed one cell per 50 sensor volumes (to avoid two cells being in the sensor at the same time), yielding the line of possible operating points.
Supplementary Figure 15 Effect of signal clipping in power spectral density around the sensor resonant frequencies.
Saturation applied to a channel carrying many sinusoids adds many other spectral components that are not easily filtered out (noise).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–15, Supplementary Table 1 and Supplementary Notes 1–4 (PDF 2553 kb)
Rights and permissions
About this article
Cite this article
Cermak, N., Olcum, S., Delgado, F. et al. High-throughput measurement of single-cell growth rates using serial microfluidic mass sensor arrays. Nat Biotechnol 34, 1052–1059 (2016). https://doi.org/10.1038/nbt.3666
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.3666
This article is cited by
-
Monitoring the mass, eigenfrequency, and quality factor of mammalian cells
Nature Communications (2024)
-
Acoustic tweezers for high-throughput single-cell analysis
Nature Protocols (2023)
-
High-throughput determination of dry mass of single bacterial cells by ultrathin membrane resonators
Communications Biology (2022)
-
Differentiating Live Versus Dead Gram-Positive and Gram-Negative Bacteria With and Without Oxidative Stress Using Buoyant Mass Measurements
Current Microbiology (2022)
-
A pipeline for malignancy and therapy agnostic assessment of cancer drug response using cell mass measurements
Communications Biology (2022)