Abstract
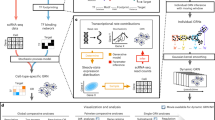
Reconstruction of the molecular pathways controlling organ development has been hampered by a lack of methods to resolve embryonic progenitor cells. Here we describe a strategy to address this problem that combines gene expression profiling of large numbers of single cells with data analysis based on diffusion maps for dimensionality reduction and network synthesis from state transition graphs. Applying the approach to hematopoietic development in the mouse embryo, we map the progression of mesoderm toward blood using single-cell gene expression analysis of 3,934 cells with blood-forming potential captured at four time points between E7.0 and E8.5. Transitions between individual cellular states are then used as input to develop a single-cell network synthesis toolkit to generate a computationally executable transcriptional regulatory network model of blood development. Several model predictions concerning the roles of Sox and Hox factors are validated experimentally. Our results demonstrate that single-cell analysis of a developing organ coupled with computational approaches can reveal the transcriptional programs that underpin organogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Shalaby, F. et al. A requirement for Flk1 in primitive and definitive hematopoiesis and vasculogenesis. Cell 89, 981–990 (1997).
Shalaby, F. et al. Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature 376, 62–66 (1995).
Guo, G. et al. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev. Cell 18, 675–685 (2010).
Yan, L. et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1131–1139 (2013).
Xue, Z. et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 500, 593–597 (2013).
Pina, C. et al. Inferring rules of lineage commitment in haematopoiesis. Nat. Cell Biol. 14, 287–294 (2012).
Moignard, V. et al. Characterization of transcriptional networks in blood stem and progenitor cells using high-throughput single-cell gene expression analysis. Nat. Cell Biol. 15, 363–372 (2013).
Swiers, G. et al. Early dynamic fate changes in haemogenic endothelium characterized at the single-cell level. Nat. Commun. 4, 2924 (2013).
Coifman, R.R. et al. Geometric diffusions as a tool for harmonic analysis and structure definition of data: diffusion maps. Proc. Natl. Acad. Sci. USA 102, 7426–7431 (2005).
Lux, C.T. et al. All primitive and definitive hematopoietic progenitor cells emerging before E10 in the mouse embryo are products of the yolk sac. Blood 111, 3435–3438 (2008).
Ding, G., Tanaka, Y., Hayashi, M., Nishikawa, S.-I. & Kataoka, H. PDGF receptor alpha+ mesoderm contributes to endothelial and hematopoietic cells in mice. Dev. Dyn. 242, 254–268 (2013).
Tanaka, Y. et al. Early ontogenic origin of the hematopoietic stem cell lineage. Proc. Natl. Acad. Sci. USA 109, 4515–4520 (2012).
Lorsbach, R.B. et al. Role of RUNX1 in adult hematopoiesis: analysis of RUNX1-IRES-GFP knock-in mice reveals differential lineage expression. Blood 103, 2522–2529 (2004).
Moignard, V., Woodhouse, S., Fisher, J. & Göttgens, B. Transcriptional hierarchies regulating early blood cell development. Blood Cells Mol. Dis. 51, 239–247 (2013).
Thiery, J.P., Acloque, H., Huang, R.Y.J. & Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell 139, 871–890 (2009).
Costa, G., Kouskoff, V. & Lacaud, G. Origin of blood cells and HSC production in the embryo. Trends Immunol. 33, 215–223 (2012).
Samokhvalov, I.M., Samokhvalova, N.I. & Nishikawa, S. Cell tracing shows the contribution of the yolk sac to adult haematopoiesis. Nature 446, 1056–1061 (2007).
Fujiwara, Y., Browne, C.P., Cunniff, K., Goff, S.C. & Orkin, S.H. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc. Natl. Acad. Sci. USA 93, 12355–12358 (1996).
Robb, L. et al. Absence of yolk sac hematopoiesis from mice with a targeted disruption of the scl gene. Proc. Natl. Acad. Sci. USA 92, 7075–7079 (1995).
Shivdasani, R.A., Mayer, E.L. & Orkin, S.H. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature 373, 432–434 (1995).
Schlaeger, T.M., Mikkola, H.K.A., Gekas, C., Helgadottir, H.B. & Orkin, S.H. Tie2Cre-mediated gene ablation defines the stem-cell leukemia gene (SCL/tal1)-dependent window during hematopoietic stem-cell development. Blood 105, 3871–3874 (2005).
Chen, M.J., Yokomizo, B.M., Zeigler, E., Dzierzak, E. & Speck, N.A. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature 457, 887–891 (2009).
North, T. et al. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development 126, 2563–2575 (1999).
Wareing, S. et al. The Flk1-Cre-mediated deletion of ETV2 defines its narrow temporal requirement during embryonic hematopoietic development. Stem Cells 30, 1521–1531 (2012).
Sumanas, S. et al. Interplay among Etsrp/ER71, Scl, and Alk8 signaling controls endothelial and myeloid cell formation. Blood 111, 4500–4510 (2008).
Krumsiek, J., Marr, C., Schroeder, T. & Theis, F.J. Hierarchical differentiation of myeloid progenitors is encoded in the transcription factor network. PLoS ONE 6, e22649 (2011).
Bonzanni, N. et al. Hard-wired heterogeneity in blood stem cells revealed using a dynamic regulatory network model. Bioinformatics 29, i80–i88 (2013).
Xu, H., Ang, Y.-S., Sevilla, A., Lemischka, I.R. & Ma'ayan, A. Construction and validation of a regulatory network for pluripotency and self-renewal of mouse embryonic stem cells. PLoS Comput. Biol. 10, e1003777 (2014).
Sánchez-Castillo, M. et al. CODEX: a next-generation sequencing experiment database for the haematopoietic and embryonic stem cell communities. Nucleic Acids Res. 43, D1117–D1123 (2015).
Fan, R. et al. Dynamic HoxB4-regulatory network during embryonic stem cell differentiation to hematopoietic cells. Blood 119, e139–e147 (2012).
Thoms, J.A. et al. ERG promotes T-acute lymphoblastic leukemia and is transcriptionally regulated in leukemic cells by a stem cell enhancer. Blood 117, 7079–7089 (2011).
Wilson, N.K. et al. The transcriptional program controlled by the stem cell leukemia gene Scl/Tal1 during early embryonic hematopoietic development. Blood 113, 5456–5465 (2009).
Wilkinson, A.C. et al. Single site-specific integration targeting coupled with embryonic stem cell differentiation provides a high-throughput alternative to in vivo enhancer analyses. Biol. Open 2, 1229–1238 (2013).
Mitjavila-Garcia, M.T. et al. Expression of CD41 on hematopoietic progenitors derived from embryonic hematopoietic cells. Development 129, 2003–2013 (2002).
Mikkola, H.K.A., Fujiwara, Y., Schlaeger, T.M., Traver, D. & Orkin, S.H. Expression of CD41 marks the initiation of definitive hematopoiesis in the mouse embryo. Blood 101, 508–516 (2003).
Kabrun, N. et al. Flk-1 expression defines a population of early embryonic hematopoietic precursors. Development 124, 2039–2048 (1997).
Gandillet, A. et al. Sox7-sustained expression alters the balance between proliferation and differentiation of hematopoietic progenitors at the onset of blood specification. Blood 114, 4813–4822 (2009).
Sachs, K., Perez, O., Pe'er, D., Lauffenburger, D.A. & Nolan, G.P. Causal protein-signaling networks derived from multiparameter single-cell data. Science 308, 523–529 (2005).
Guo, G. et al. Mapping cellular hierarchy by single-cell analysis of the cell surface repertoire. Cell Stem Cell 13, 492–505 (2013).
Bailey, N.T.J. Statistical Methods in Biology 1995, 255 (Cambridge University Press, 1995).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Pimanda, J.E. et al. Gata2, Fli1, and Scl form a recursively wired gene-regulatory circuit during early hematopoietic development. Proc. Natl. Acad. Sci. USA 104, 17692–17697 (2007).
Kataoka, H. et al. Etv2/ER71 induces vascular mesoderm from Flk1+PDGFRα+ primitive mesoderm. Blood 118, 6975–6986 (2011).
Loughran, S.J. et al. The transcription factor Erg is essential for definitive hematopoiesis and the function of adult hematopoietic stem cells. Nat. Immunol. 9, 810–819 (2008).
Taoudi, S. et al. ERG dependence distinguishes developmental control of hematopoietic stem cell maintenance from hematopoietic specification. Genes Dev. 25, 251–262 (2011).
Sacilotto, N. et al. Analysis of Dll4 regulation reveals a combinatorial role for Sox and Notch in arterial development. Proc. Natl. Acad. Sci. USA 110, 11893–11898 (2013).
Kyba, M., Perlingeiro, R.C.R. & Daley, G.Q. HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell 109, 29–37 (2002).
Tischler, J. & Surani, M.A. Investigating transcriptional states at single-cell-resolution. Curr. Opin. Biotechnol. 24, 69–78 (2013).
Tang, F., Lao, K. & Surani, M.A. Development and applications of single-cell transcriptome analysis. Nat. Methods 8, S6–S11 (2011).
Downs, K.M. & Davies, T. Staging of gastrulating mouse embryos by morphological landmarks in the dissecting microscope. Development 118, 1255–1266 (1993).
Ståhlberg, A. et al. Defining cell populations with single-cell gene expression profiling: correlations and identification of astrocyte subpopulations. Nucleic Acids Res. 39, e24 (2011).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Van der Maaten, L. & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Amir, E.-A.D. et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 31, 545–552 (2013).
Koksal, A.S. et al. Synthesis of biological models from mutation experiments. POPL '13 Proc. 40th Annu. ACM SIGPLAN-SIGACT Symp. Princ. Program. Lang 48, 469–482 (2013).
Garg, A., Di Cara, A., Xenarios, I., Mendoza, L. & De Micheli, G. Synchronous versus asynchronous modeling of gene regulatory networks. Bioinformatics 24, 1917–1925 (2008).
Magin, T.M., McWhir, J. & Melton, D.W. A new mouse embryonic stem cell line with good germ line contribution and gene targeting frequency. Nucleic Acids Res. 20, 3795–3796 (1992).
Acknowledgements
We thank J. Downing (St. Jude Children's Research Hospital, Memphis, TN, USA) for the Runx1-ires-GFP mouse. Research in the authors' laboratory is supported by the Medical Research Council, Biotechnology and Biological Sciences Research Council, Leukaemia and Lymphoma Research, the Leukemia and Lymphoma Society, Microsoft Research and core support grants by the Wellcome Trust to the Cambridge Institute for Medical Research and Wellcome Trust - MRC Cambridge Stem Cell Institute. V.M. is supported by a Medical Research Council Studentship and Centenary Award and S.W. by a Microsoft Research PhD Scholarship.
Author information
Authors and Affiliations
Contributions
V.M., A.J.L., Y.T., A.C.W. and I.C.M. performed experiments and analyzed data. S.W., L.H., F.B. and N.P. developed computational tools. W.J. and E.D. analyzed data. S.-I.N., V.K., F.J.T., J.F. and B.G. conceived the study. V.M., S.W., J.F. and B.G. wrote the paper with help from all co-authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Fluorescence activated cell sorting of cells with blood potential.
(a) Gating strategy for Flk1+ cells at PS, NP and HF stages, and Runx1-ires-GFP+ cells (4SG) or Flk1+Runx1-ires-GFP− cells (4SFG−) at the 4S stage. (b) Total numbers of cells per embryo at each stage, estimated from FACS data. Each point represents one embryo and lines indicate the median.
Supplementary Figure 2 Development is asynchronous.
(a) PCA of the 3,934 cells, coloured retrospectively according to the stage from which they were. Blue, PS; green, NP; orange, HF; red, 4SG; purple, 4SFG−. (b) For each stage, the cells from different embryos are shown on the PCA as different colours (cells from other stages shown in grey). (c) PCA coloured according to the clusters cells belong to in Figure 1e. Green, I; Blue, II; Pink, III. (d) For each embryo, the percentage of cells in clusters 1, 2 and 3 was calculated. The mean and s.d. were then calculated for each cluster in each stage. Number of embryos shown in Fig. 1c.
Supplementary Figure 3 Flk1+Runx1+ cells are found in all anatomical stages.
Principal component analysis coloured according to stage of sorting (top) and whether cells express both Flk1 and Runx1 at the gene expression level (bottom). Cells not expressing both genes shown in grey. Blue, PS; green, NP; orange, HF; red, 4SG; purple, 4SFG−.
Supplementary Figure 4 RNA sequencing of populations of 50 cells.
Schematic of early embryonic development of the mesodermal lineage. The cardiac, smooth muscle, endothelial and blood lineages all diverge from Flk1+ mesoderm, while somatic muscle diverges earlier. (a-f) Populations of 50 cells were sorted from 5 embryos each at PS, NP and HF stages. FPKM values for three key genes are shown for each of the lineage decisions in the schematic. Points are the mean and s.d. of the 5 replicates.
Supplementary Figure 5 Diffusion plots.
Diffusion plots showing expression patterns of additional genes not shown in Figure 2.
Supplementary Figure 6 Diffusion plots highlighting each stage.
Diffusion plots were coloured according to embryonic stage. Other cells shown in grey. Blue, PS; green, NP; orange, HF; red, 4SG; purple, 4SFG-.
Supplementary Figure 7 Comparison of multivariate analysis techniques.
The data were plotted using diffusion maps, principal component analysis, independent component analysis and t-SNE. Blue, PS; green, NP; orange, HF; red, 4SG; purple, 4SFG-.
Supplementary Figure 8 Some states occur in multiple cells.
State transition graph coloured by the number of times each binary state occurs in the 3,934 expression profiles. Grey, occurs once; blue, occurs twice; red, occurs more than twice.
Supplementary Figure 9 Populations mask variation and asynchrony.
Pools of 20 cells were simulated from single cell data and projected onto the principal component analysis for all 3,934 cells. Each plot shows one embryonic stage and the simulated pools of 20 cells for that population in black (normalized in the same way as single cells). Bottom right hand plot shows the head fold stage and simulated pools as well as actual pools of 20 cells sorted from embryos (pink).
Supplementary Figure 10 Analysis of transcription factor binding sites in network gene loci.
(a) Motifs for DNA-binding TFs in the core network. Note that Eto2 and Lmo2 do not bind directly to the DNA, and the binding site of the Notch partner Rbpj was used for Notch1 as described previously. (b) To search for evidence that predicted regulatory interactions are direct, all haematopoietic TF ChIP-seq data in Codex2 were searched for peaks within the gene body or 50 kb up- and down-stream of the 20 network genes. Overlapping peaks were merged to identify putative regulatory sequences. Different colours indicate peaks from different experiments. A transcription factor binding site search (TFBS) was performed using TFBSsearch3 on the mouse genomic sequence of all regions to identify the consensus binding sites of the network TFs. Evolutionary conservation of TF binding motifs is a well-recognised feature of truly functional binding sites4. To identify conserved binding sites, PhyloP scores were calculated for each motif across 60 species using the tool provided by the UCSC Genome Browser. Evolutionarily conserved motifs previously validated in functional assays by us and other were characterised by PhyloP scores greater than 1, which was therefore used as the threshold. Results are summarised in Supplementary Table 2.
Supplementary Figure 11 The Erg+85 enhancer is active during haematopoietic development
(a) UCSC Genome Browser tracks of re-analysed ChIP-Seq data for HoxB45 indicates that HoxB4 binds to the Erg+85 enhancer (shown in grey) during a 26-day differentiation time course that produces HSCs from ES cells, with HSCs able to contribute to long-term engraftment in recipient mice. Binding is absent at day 6, but becomes apparent by day 16. (b) Flow cytometric analysis of Flk1 and CD41 expression across a time-course of EB differentiation from Figure 4B indicates that different clones have similar kinetics of differentiation, with Flk1+ cells giving rise to Flk1+CD41+ and then Flk1−CD41+ cells. (c) Flow cytometric analysis of YFP expression in all live cells across a time-course of EB differentiation from Figure 4b illustrates reduction of YFP positive cells in ES cell clones where YFP expression is driven by mutant enhancer constructs. In (b) and (c), a representative experiment is shown from three biological repeats of 2-3 clones per construct.
Supplementary Figure 12 Partial correlation analysis.
(a) Hierarchical clustering of partial correlation coefficients between pairs of transcription factors for all 3,934 expression profiles. Pairwise coefficients were calculated while controlling for the effect of all other transcription factors. Erg does not correlate with Sox or Hox factors. (b) Network diagram showing putative undirected activating (red) and inhibiting (blue) relationships suggested by significant correlations (p-value < 1e-10).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–12, Supplementary Tables 1–4, 6 and Supplementary Note (PDF 14079 kb)
Supplementary Table 5
Summary of in silico perturbations (XLSX 89 kb)
Supplementary Table 7
Raw Ct values and normalised dCt values for the 3,934 cells presented in this paper (XLSX 3322 kb)
Supplementary Table 8
FPKM values for RNAseq of 50 cell populations (XLSX 5225 kb)
Supplementary Table 9
Initial and final states used in network synthesis (XLSX 40 kb)
Supplementary Table 10
Binary gene expression data (XLSX 204 kb)
Supplementary Table 11
List of cells with equivalent binary expression states (XLSX 46 kb)
Rights and permissions
About this article
Cite this article
Moignard, V., Woodhouse, S., Haghverdi, L. et al. Decoding the regulatory network of early blood development from single-cell gene expression measurements. Nat Biotechnol 33, 269–276 (2015). https://doi.org/10.1038/nbt.3154
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.3154
This article is cited by
-
Inference on autoregulation in gene expression with variance-to-mean ratio
Journal of Mathematical Biology (2023)
-
NetAct: a computational platform to construct core transcription factor regulatory networks using gene activity
Genome Biology (2022)
-
Chemotherapy induces canalization of cell state in childhood B-cell precursor acute lymphoblastic leukemia
Nature Cancer (2021)
-
Mapping a mammalian adult adrenal gland hierarchy across species by microwell-seq
Cell Regeneration (2020)
-
Analysis on gene modular network reveals morphogen-directed development robustness in Drosophila
Cell Discovery (2020)