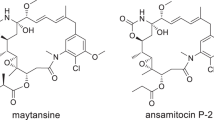
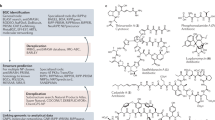
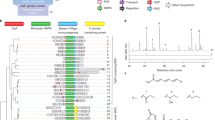
Abstract
Microbially derived natural products are major sources of antibiotics and other medicines, but discovering new antibiotic scaffolds and increasing the chemical diversity of existing ones are formidable challenges. We have designed a screen to exploit the self-protection mechanism of antibiotic producers to enrich microbial libraries for producers of selected antibiotic scaffolds. Using resistance as a discriminating criterion we increased the discovery rate of producers of both glycopeptide and ansamycin antibacterial compounds by several orders of magnitude in comparison with historical hit rates. Applying a phylogeny-based screening filter for biosynthetic genes enabled the binning of producers of distinct scaffolds and resulted in the discovery of a glycopeptide antibacterial compound, pekiskomycin, with an unusual peptide scaffold. This strategy provides a means to readily sample the chemical diversity available in microbes and offers an efficient strategy for rapid discovery of microbial natural products and their associated biosynthetic enzymes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Wright, G.D. Antibiotics: a new hope. Chem. Biol. 19, 3–10 (2012).
Anonymous. A call to arms. Nat. Rev. Drug Discov. 6, 8–12 (2007).
Cooper, M.A. & Shlaes, D. Fix the antibiotics pipeline. Nature 472, 32 (2011).
Reddy, B.V.B. et al. Natural product biosynthetic gene diversity in geographically distinct soil microbiomes. Appl. Environ. Microbiol. 78, 3744–3752 (2012).
Nett, M., Ikeda, H. & Moore, B.S. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat. Prod. Rep. 26, 1362–1384 (2009).
Lamb, S.S. & Wright, G.D. Accessorizing natural products: adding to nature's toolbox. Proc. Natl. Acad. Sci. USA 102, 519–520 (2005).
Baltz, R.H. Marcel Faber Roundtable: is our antibiotic pipeline unproductive because of starvation, constipation or lack of inspiration? J. Ind. Microbiol. Biotechnol. 33, 507–513 (2006).
Thaker, M.N. & Wright, G.D. Opportunities for synthetic biology in antibiotics: expanding glycopeptide chemical diversity. ACS Synth. Biol. doi:10.1021/sb300092n (17 December 2012).
Cundliffe, E. & Demain, A.L. Avoidance of suicide in antibiotic-producing microbes. J. Ind. Microbiol. Biotechnol. 37, 643–672 (2010).
Wright, G.D. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 5, 175–186 (2007).
Courvalin, P. Vancomycin resistance in Gram-positive cocci. Clin. Infect. Dis. 42, S25–S34 (2006).
Chiu, H.T. et al. Molecular cloning and sequence analysis of the complestatin biosynthetic gene cluster. Proc. Natl. Acad. Sci. USA 98, 8548–8553 (2001).
Lanoot, B. et al. BOX-PCR fingerprinting as a powerful tool to reveal synonymous names in the genus Streptomyces. Emended descriptions are proposed for the species Streptomyces cinereorectus, S. fradiae, S. tricolor, S. colombiensis, S. filamentosus, S. vinaceus and S. phaeopurpureus. Syst. Appl. Microbiol. 27, 84–92 (2004).
Thaker, M.N. et al. Biosynthetic gene cluster and antimicrobial activity of the elfamycin antibiotic factumycin. Med. Chem. Commun. 3, 1020–1026 (2012).
Nagarajan, R. et al. M43 antibiotics: methylated vancomycins and unrearranged CDP-I analogs. J. Am. Chem. Soc. 110, 7896–7897 (1988).
Yim, G., Wang, H.H. & Davies, J. Antibiotics as signalling molecules. Phil. Trans. R. Soc. Lond. B 362, 1195–1200 (2007).
Pootoolal, J. et al. Assembling the glycopeptide antibiotic scaffold: The biosynthesis of A47934 from Streptomyces toyocaensis NRRL15009. Proc. Natl. Acad. Sci. USA 99, 8962–8967 (2002).
Skelton, N.J., Williams, D.H., Monday, R.A. & Ruddock, J.C. Structure elucidation of the novel glycopeptide antibiotic UK-68,597. J. Org. Chem. 55, 3718–3723 (1990).
Banik, J.J. & Brady, S.F. Cloning and characterization of new glycopeptide gene clusters found in an environmental DNA megalibrary. Proc. Natl. Acad. Sci. USA 105, 17273–17277 (2008).
Banik, J.J., Craig, J.W., Calle, P.Y. & Brady, S.F. Tailoring enzyme-rich environmental DNA clones: a source of enzymes for generating libraries of unnatural natural products. J. Am. Chem. Soc. 132, 15661–15670 (2010).
Bibikova, M.V., Ivanitskaia, L.P. & Singal, E.M. Antibiotiki Directed screening of aminoglycoside antibiotic producers on selective media with gentamycin. (Original in Russian.) 26, 488–492 (1981).
Ivanitskaia, L.P., Bibikova, M.V., Gromova, M.N., Zhdanovich Iu, V. & Istratov, E.N. Antibiotiki Use of selective media with lincomycin for the directed screening of antibiotic producers. (Original in Russian.) 26, 83–86 (1981).
Hong, H.J., Hutchings, M.I. & Buttner, M.J. Vancomycin resistance VanS/VanR two-component systems. Adv. Exp. Med. Biol. 631, 200–213 (2008).
Koteva, K. et al. A vancomycin photoprobe identifies the histidine kinase VanSsc as a vancomycin receptor. Nat. Chem. Biol. 6, 327–329 (2010).
Berdy, J. Bioactive microbial metabolites. J. Antibiot. (Tokyo) 58, 1–26 (2005).
Hosaka, T. et al. Antibacterial discovery in actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nat. Biotechnol. 27, 462–464 (2009).
Martín, J.-F. & Liras, P. Engineering of regulatory cascades and networks controlling antibiotic biosynthesis in Streptomyces. Curr. Opin. Microbiol. 13, 263–273 (2010).
D'Costa, V.M., McGrann, K.M., Hughes, D.W. & Wright, G.D. Sampling the antibiotic resistome. Science 311, 374–377 (2006).
Hayakawa, M. & Nonomura, H. Humic acid-vitamin agar, a new medium for the selective isolation of soil actinomycetes. J. Ferment.Technol. 65, 501–509 (1987).
He, W., Wu, L., Gao, Q., Du, Y. & Wang, Y. Identification of AHBA biosynthetic genes related to geldanamycin biosynthesis in Streptomyces hygroscopicus 17997. Curr. Microbiol. 52, 197–203 (2006).
Edgar, R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Folena-Wasserman, G., Sitrin, R.D., Chapin, F. & Snader, K.M. Affinity chromatography of glycopeptide antibiotics. J. Chromatogr. A 392, 225–238 (1987).
Li, H. Exploring single-sample SNP and INDEL calling with whole-genome de novo assembly. Bioinformatics 28, 1838–1844 (2012).
Chevreux, B., Wetter, T. & Suhai, S. in Computer Science and Biology: Proceedings of the German Conference on Bioinformatics, Hannover, Germany, 4–6 October, pp. 45–56 (GCB, 1999).
Altschul, S.F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Medema, M.H. et al. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 39, W339–W346 (2011).
Acknowledgements
We are grateful to X.D. Wang for isolation of actinomycetes, K. Koteva for sharing expertise in GPA purification, C. King for genome sequencing and C. Quinn, TA Instruments for help with ITC data. This research was funded by a Canadian Institutes of Health Research (CIHR) Grant MT-14981, Natural Sciences and Engineering Research Council Grant (237480) and by a Canada Research Chair in Antibiotic Biochemistry (G.D.W.).
Author information
Authors and Affiliations
Contributions
M.N.T. designed the experiments, isolated genomic DNA, designed PCR primers, standardized PCR conditions, and carried out BOX PCRs and other PCRs, performed isolation and purification of GPAs, performed in silico genome analysis, determined MIC values and wrote the manuscript. W.W. purified the GPAs, carried out NMR experiments and their analysis, and elucidated the structures. P.S. designed and carried out experiments for the resistance-based discovery of ansamycins. N.W. performed genome assemblies, created phylogenies and bioinformatic analyses, and submitted sequences. A.M.K. designed primers and isolated genomic DNA. R.M. isolated genomic DNA, performed 16S rDNA PCRs and fingerprinting PCRs. G.D.W. developed the concept, designed the experiments and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–21 and Supplementary Tables 1–9 (PDF 4376 kb)
Rights and permissions
About this article
Cite this article
Thaker, M., Wang, W., Spanogiannopoulos, P. et al. Identifying producers of antibacterial compounds by screening for antibiotic resistance. Nat Biotechnol 31, 922–927 (2013). https://doi.org/10.1038/nbt.2685
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.2685
This article is cited by
-
Diverse secondary metabolites are expressed in particle-associated and free-living microorganisms of the permanently anoxic Cariaco Basin
Nature Communications (2023)
-
Genome-based classification of Streptomyces anatolicus sp. nov., an actinobacterium with antimicrobial and cytotoxic activities, and reclassification of Streptomyces nashvillensis as a later heterotypic synonym of Streptomyces tanashiensis
Antonie van Leeuwenhoek (2023)
-
Characterization of the soil resistome and mobilome in Namib Desert soils
International Microbiology (2023)
-
Newest perspectives of glycopeptide antibiotics: biosynthetic cascades, novel derivatives, and new appealing antimicrobial applications
World Journal of Microbiology and Biotechnology (2023)
-
Integrated genomics and proteomics analysis of Paenibacillus peoriae IBSD35 and insights into its antimicrobial characteristics
Scientific Reports (2022)