Abstract
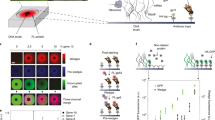
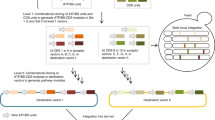
Low-cost, high-throughput gene synthesis and precise control of protein expression are of critical importance to synthetic biology and biotechnology1,2,3. Here we describe the development of an on-chip gene synthesis technology, which integrates on a single microchip the synthesis of DNA oligonucleotides using inkjet printing, isothermal oligonucleotide amplification and parallel gene assembly. Use of a mismatch-specific endonuclease for error correction results in an error rate of ∼0.19 errors per kb. We applied this approach to synthesize pools of thousands of codon-usage variants of lacZα and 74 challenging Drosophila protein antigens, which were then screened for expression in Escherichia coli. In one round of synthesis and screening, we obtained DNA sequences that were expressed at a wide range of levels, from zero to almost 60% of the total cell protein mass. This technology may facilitate systematic investigation of the molecular mechanisms of protein translation and the design, construction and evolution of macromolecular machines, metabolic networks and synthetic cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Tian, J., Ma, K. & Saaem, I. Advancing high-throughput gene synthesis technology. Mol. Biosyst. 5, 714–722 (2009).
Carr, P.A. & Church, G.M. Genome engineering. Nat. Biotechnol. 27, 1151–1162 (2009).
Welch, M., Villalobos, A., Gustafsson, C. & Minshull, J. You're one in a googol: optimizing genes for protein expression. J. R. Soc. Interface 6 Suppl 4, S467–S476 (2009).
Patwardhan, R.P. et al. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nat. Biotechnol. 27, 1173–1175 (2009).
Pfleger, B.F., Pitera, D.J., Smolke, C.D. & Keasling, J.D. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat. Biotechnol. 24, 1027–1032 (2006).
Salis, H.M., Mirsky, E.A. & Voigt, C.A. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 27, 946–950 (2009).
Isaacs, F.J. et al. Engineered riboregulators enable post-transcriptional control of gene expression. Nat. Biotechnol. 22, 841–847 (2004).
Kudla, G., Murray, A.W., Tollervey, D. & Plotkin, J.B. Coding-sequence determinants of gene expression in Escherichia coli. Science 324, 255–258 (2009).
Coleman, J.R. et al. Virus attenuation by genome-scale changes in codon pair bias. Science 320, 1784–1787 (2008).
Dueber, J.E. et al. Synthetic protein scaffolds provide modular control over metabolic flux. Nat. Biotechnol. 27, 753–759 (2009).
Basu, S., Gerchman, Y., Collins, C.H., Arnold, F.H. & Weiss, R. A synthetic multicellular system for programmed pattern formation. Nature 434, 1130–1134 (2005).
Danino, T., Mondragon-Palomino, O., Tsimring, L. & Hasty, J. A synchronized quorum of genetic clocks. Nature 463, 326–330 (2010).
Ellis, T., Wang, X. & Collins, J.J. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat. Biotechnol. 27, 465–471 (2009).
Tabor, J.J. et al. A synthetic genetic edge detection program. Cell 137, 1272–1281 (2009).
Gibson, D.G. et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science 319, 1215–1220 (2008).
Tian, J. et al. Accurate multiplex gene synthesis from programmable DNA microchips. Nature 432, 1050–1054 (2004).
Zhou, X. et al. Microfluidic PicoArray synthesis of oligodeoxynucleotides and simultaneous assembling of multiple DNA sequences. Nucleic Acids Res. 32, 5409–5417 (2004).
Richmond, K.E. et al. Amplification and assembly of chip-eluted DNA (AACED): a method for high-throughput gene synthesis. Nucleic Acids Res. 32, 5011–5018 (2004).
Borovkov, A.Y. et al. High-quality gene assembly directly from unpurified mixtures of microarray-synthesized oligonucleotides. Nucleic Acids Res. 38, e180 (2010).
Kosuri, S. et al. Scalable gene synthesis by selective amplification of DNA pools from high-fidelity microchips. Nat. Biotechnol. 28, 1295–1299 (2010).
Saaem, I., Ma, K., Marchi, A., LaBean, T. & Tian, J. In situ synthesis of DNA microarray on functionalized cyclic olefin copolymer substrate. ACS Applied Materials & Interfaces 2, 491–497 (2010).
Ma, K.-S., Reza, F., Saaem, I. & Tian, J. Versatile surface functionalization of cyclic olefin copolymer (COC) with sputtered SiO2 thin film for potential BioMEMS applications. J. Mater. Chem. 19, 7914–7920 (2009).
Huang, M.C., Ye, H., Kuan, Y.K., Li, M.H. & Ying, J.Y. Integrated two-step gene synthesis in a microfluidic device. Lab Chip 9, 276–285 (2009).
Qiu, P. et al. Mutation detection using Surveyor nuclease. Biotechniques 36, 702–707 (2004).
The ENCODE Project Consortium. The E.N.C.O.D.E. (ENCyclopedia Of DNA Elements) Project. Science 306, 636–640 (2004).
Nakamura, Y., Gojobori, T. & Ikemura, T. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 28, 292 (2000).
Quan, J. & Tian, J. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS ONE 4, e6441 (2009).
Sharp, P.M. & Li, W.H. The codon Adaptation Index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15, 1281–1295 (1987).
Lamprecht, M.R., Sabatini, D.M. & Carpenter, A.E. CellProfiler: free, versatile software for automated biological image analysis. Biotechniques 42, 71–75 (2007).
Quan, J. & Tian, J. Circular polymerase extension cloning for high-throughput cloning of complex and combinatorial DNA libraries. Nat. Protoc. 6, 242–251 (2011).
Acknowledgements
We thank Z. Chen, K. Ma and Q. Wang for excellent technical contributions. The project was supported by the Beckman foundation, The Hartwell Foundation, and a Coulter-Duke translational partnership award to J.T.
Author information
Authors and Affiliations
Contributions
J.Q. performed gene library synthesis, expression screen and protein purification experiments. I.S. performed on-chip oligo and gene synthesis experiments. N.T. and J.Q. performed colony imaging and image analysis. S.M., I.S. and J.Q. performed error correction experiments. N.N. and K.P.W. provided wild-type transcription factor sequences and clones. H.G. and J.T. designed gene library sequences. J.T. designed overall strategy and supervised the project. All authors contributed to the manuscript preparation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–3, Supplementary Figs. 1–4, Supplementary Sequences and Supplementary Note (PDF 698 kb)
Rights and permissions
About this article
Cite this article
Quan, J., Saaem, I., Tang, N. et al. Parallel on-chip gene synthesis and application to optimization of protein expression. Nat Biotechnol 29, 449–452 (2011). https://doi.org/10.1038/nbt.1847
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.1847
This article is cited by
-
An open-source, 3D printed inkjet DNA synthesizer
Scientific Reports (2024)
-
Purification of multiplex oligonucleotide libraries by synthesis and selection
Nature Biotechnology (2022)
-
Glycan chip based on structure-switchable DNA linker for on-chip biosynthesis of cancer-associated complex glycans
Nature Communications (2021)
-
Codon optimization with deep learning to enhance protein expression
Scientific Reports (2020)
-
An oligonucleotide synthesizer based on a microreactor chip and an inkjet printer
Scientific Reports (2019)