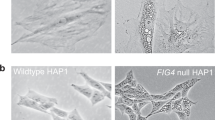
Abstract
Large collections of knockout organisms facilitate the elucidation of gene functions. Here we used retroviral insertion or homologous recombination to disrupt 472 genes encoding secreted and membrane proteins in mice, providing a resource for studying a large fraction of this important class of drug target. The knockout mice were subjected to a systematic phenotypic screen designed to uncover alterations in embryonic development, metabolism, the immune system, the nervous system and the cardiovascular system. The majority of knockout lines exhibited altered phenotypes in at least one of these therapeutic areas. To our knowledge, a comprehensive phenotypic assessment of a large number of mouse mutants generated by a gene-specific approach has not been described previously.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Clamp, M. et al. Distinguishing protein-coding and noncoding genes in the human genome. Proc. Natl. Acad. Sci. USA 104, 19428–19433 (2007).
Austin, C.P. et al. The knockout mouse project. Nat. Genet. 36, 921–924 (2004).
Friedel, R.H., Seisenberger, C., Kaloff, C. & Wurst, W. EUCOMM–the European conditional mouse mutagenesis program. Brief. Funct. Genomics Proteomics 6, 180–185 (2007).
Mouse Genome Sequencing Consortium & Waterston, R.H. et al. Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 (2002).
Zambrowicz, B.P. & Sands, A.T. Knockouts model the 100 best-selling drugs–will they model the next 100? Nat. Rev. Drug Discov. 2, 38–51 (2003).
Clark, H.F. et al. The secreted protein discovery initiative (SPDI), a large-scale effort to identify novel human secreted and transmembrane proteins: a bioinformatics assessment. Genome Res. 13, 2265–2270 (2003).
Zambrowicz, B.P. et al. Disruption and sequence identification of 2,000 genes in mouse embryonic stem cells. Nature 392, 608–611 (1998).
Beltrandelrio, H. et al. Saturation Screening of the Druggable Mammalian Genome. in Model Organisms in Drug Discovery (eds. Carroll, P.M. & Fitzgerald, K.) 251–278, (John Wiley & Sons, Chichester, West Sussex, England, 2003).
Brommage, R. Validation and calibration of DEXA body composition in mice. Am. J. Physiol. Endocrinol. Metab. 285, E454–E459 (2003).
Scott, H.S. et al. Insertion of beta-satellite repeats identifies a transmembrane protease causing both congenital and childhood onset autosomal recessive deafness. Nat. Genet. 27, 59–63 (2001).
Guipponi, M., Antonarakis, S.E. & Scott, H.S. TMPRSS3, a type II transmembrane serine protease mutated in non-syndromic autosomal recessive deafness. Front. Biosci. 13, 1557–1567 (2008).
Ben-Yosef, T. et al. Claudin 14 knockout mice, a model for autosomal recessive deafness DFNB29, are deaf due to cochlear hair cell degeneration. Hum. Mol. Genet. 12, 2049–2061 (2003).
Friedman, L.M., Dror, A.A. & Avraham, K.B. Mouse models to study inner ear development and hereditary hearing loss. Int. J. Dev. Biol. 51, 609–631 (2007).
Fan, B. et al. Hepatocyte growth factor activator inhibitor-1 (HAI-1) is essential for the integrity of basement membranes in the developing placental labyrinth. Dev. Biol. 303, 222–230 (2007).
Yan, M. & Plowman, G.D. Delta-like 4/Notch signaling and its therapeutic implications. Clin. Cancer Res. 13, 7243–7246 (2007).
Wilson, L. et al. Random mutagenesis of proximal mouse chromosome 5 uncovers predominantly embryonic lethal mutations. Genome Res. 15, 1095–1105 (2005).
Junge, H.J. et al. TSPAN12 regulates retinal vascular development by promoting Norrin- but not Wnt-induced FZD4/β-catenin signaling. Cell 139, 299–311 (2009).
Beigneux, A.P. et al. Glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 plays a critical role in the lipolytic processing of chylomicrons. Cell Metab. 5, 279–291 (2007).
Desai, U. et al. Lipid-lowering effects of anti-angiopoietin-like 4 antibody recapitulate the lipid phenotype found in angiopoietin-like 4 knockout mice. Proc. Natl. Acad. Sci. USA 104, 11766–11771 (2007).
Savelieva, K.V. et al. Learning and memory impairment in Eph receptor A6 knockout mice. Neurosci. Lett. 438, 205–209 (2008).
Holst, C.R. et al. Secreted sulfatases Sulf1 and Sulf2 have overlapping yet essential roles in mouse neonatal survival. PLoS ONE 2, e575 (2007).
Colonna, M., Samaridis, J. & Angman, L. Molecular characterization of two novel C-type lectin-like receptors, one of which is selectively expressed in human dendritic cells. Eur. J. Immunol. 30, 697–704 (2000).
Turner, M. et al. Perinatal lethality and blocked B-cell development in mice lacking the tyrosine kinase Syk. Nature 378, 298–302 (1995).
Cheng, A.M. et al. Syk tyrosine kinase required for mouse viability and B-cell development. Nature 378, 303–306 (1995).
Suzuki-Inoue, K. et al. A novel Syk-dependent mechanism of platelet activation by the C-type lectin receptor CLEC-2. Blood 107, 542–549 (2006).
May, F. et al. CLEC-2 is an essential platelet activating receptor in hemostasis and thrombosis. Blood 114, 3464–3472 (2009).
Cordes, S.P. N-ethyl-N-nitrosourea mutagenesis: boarding the mouse mutant express. Microbiol. Mol. Biol. Rev. 69, 426–439 (2005).
Su, A.I. et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA 101, 6062–6067 (2004).
The Eumorphia Consortium EMPReSS: standardized phenotype screens for functional annotation of the mouse genome. Nat. Genet. 37, 1155 (2005).
Morgan, H. et al. EuroPhenome: a repository for high-throughput mouse phenotyping data. Nucleic Acids Res. 38, D577–D585 (2010).
Zambrowicz, B.P., Holt, K.H., Walke, D.W., Kirkpatrick, L.L. & Eberhart, D.E. Generation of transgenic animals. in Target Validation in Drug Discovery (eds. Metcalf, B.W. & Dillon, S.) 3–26, (Academic Press, Burlington, Massachusetts, USA, 2007).
Friddle, C.J. et al. High-throughput mouse knockouts provide a functional analysis of the genome. Cold Spring Harb. Symp. Quant. Biol. 68, 311–315 (2003).
Pogorelov, V.M., Baker, K.B., Malbari, M.M., Lanthorn, T.H. & Savelieva, K.V. A standardized behavioral test battery to identify and validate targets for neuropsychiatric diseases and pain. in Experimental Animal Models in Neurobehavioral Research (eds. Kalueff, A.V. & LaPorte, J.L.) 17–45 (Laboratory of Clinical Science, Nat. Inst. of Mental Health, Bethesda, Maryland, USA, 2008).
Brommage, R. et al. High-throughput screening of mouse knockout lines identifies true lean and obese phenotypes. Obesity (Silver Spring) 16, 2362–2367 (2008).
Zambrowicz, B.P. et al. Wnk1 kinase deficiency lowers blood pressure in mice: a gene-trap screen to identify potential targets for therapeutic intervention. Proc. Natl. Acad. Sci. USA 100, 14109–14114 (2003).
Abuin, A., Hansen, G.M. & Zambrowicz, B. Gene trap mutagenesis. in Conditional Mutagenesis: An Approach to Disease Models (eds. Feil, R. & Metzger, D.) 129–147, (Springer, 2007).
Wattler, S., Kelly, M. & Nehls, M. Construction of gene targeting vectors from lambda KOS genomic libraries. Biotechniques 26, 1150–1156, 1158, 1160 (1999).
Acknowledgements
We thank J. Brennan, S. Bunting, L. Corson, P. Fielder, E. Filvaroff, D. French, J. Junutula, F. Peale, H. Phillips, M. Rohrer, H. Stern, J. Zha, R. Watts, B. Wolf and scientists in the Genentech Immunology Department for critical review of the knockout phenotypes and E. Bierwagen and D. Wan for the bioinformatics infrastructures used to track phenotypic calls. We thank J. Mitchell for analysis and plotting of histograms depicting phenotypic ranges. We also thank M. Tessier-Lavigne, F. Bazan, M. Kong-Beltran, J. Theunissen, S. Warming and Z. Zhang for critical reading of the manuscript.
Author information
Authors and Affiliations
Contributions
F.J.d.S., T.T., A.P. and B.P.Z. designed the project, analyzed data and wrote the manuscript. F.M. and P.G. designed experiments and analyzed data. J.T. and N.G. contributed to the identification of murine orthologs. K.H.H. and N.G. contributed to the design and verification of targeting strategies. T.O., K.A.P., D.S.R., G.M.H., A.A., D.E.E., K.H.H. and B.P.Z. designed, performed and supervised the knockout generation and phenotype screen. T.T., L.L. and W.F. contributed to the statistical analysis and making the phenotype calls. J.T. and Y.L. implemented the database for public access. L.L. compiled the adult tissue calls. L.P. and W.Y. performed the embryonic in situ hybridization screen. W.Y.L., F.M., D.G. and M.S. performed the follow-up characterization of the Clec1b mutant line.
Corresponding author
Ethics declarations
Competing interests
T.T., L.L., J.T., Y.L., W.Y.L., F.M., D.G., M.S., L.P., W.Y., W.F., N.G., P.G., A.P. and F.J.d.S. are employees of Genentech, a wholly owned subsidiary of Hoffmann-La Roche. T.O., K.A.P., D.S.R., G.M.H., A.A., D.E.E., K.H.H. and B.P.Z. are employees and shareholders of Lexicon Pharmaceuticals.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1,3,4,5,8 and Supplementary Figs. 1 and 2 (PDF 807 kb)
Supplementary Table 2
Gene symbol (human) (XLS 607 kb)
Supplementary Table 7
Calls from quantitative analysis (XLS 391 kb)
Supplementary Table 6
Calls from non-quantitative assays (XLS 49 kb)
Rights and permissions
About this article
Cite this article
Tang, T., Li, L., Tang, J. et al. A mouse knockout library for secreted and transmembrane proteins. Nat Biotechnol 28, 749–755 (2010). https://doi.org/10.1038/nbt.1644
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.1644
This article is cited by
-
Disruption of sugar nucleotide clearance is a therapeutic vulnerability of cancer cells
Nature (2023)
-
IGSF11 and VISTA: a pair of promising immune checkpoints in tumor immunotherapy
Biomarker Research (2022)
-
Structural variants and tandem repeats in the founder individuals of four F2 pig crosses and implications to F2 GWAS results
BMC Genomics (2022)
-
Transmembrane protein 108 inhibits the proliferation and myelination of oligodendrocyte lineage cells in the corpus callosum
Molecular Brain (2022)
-
Additional findings of tibial dysplasia in a male with orofaciodigital syndrome type XVI
Human Genome Variation (2022)