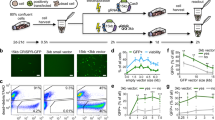
Abstract
RNA interference (RNAi) induced by short interfering RNA (siRNA) allows for discovery research and large-scale screening1,2,3,4,5; however, owing to their size and anionic charge, siRNAs do not readily enter cells4,5. Current approaches do not deliver siRNAs into a high percentage of primary cells without cytotoxicity. Here we report an efficient siRNA delivery approach that uses a peptide transduction domain–double-stranded RNA-binding domain (PTD-DRBD) fusion protein. DRBDs bind to siRNAs with high avidity, masking the siRNA's negative charge and allowing PTD-mediated cellular uptake. PTD-DRBD–delivered siRNA induced rapid RNAi in a large percentage of various primary and transformed cells, including T cells, human umbilical vein endothelial cells and human embryonic stem cells. We observed no cytotoxicity, minimal off-target transcriptional changes and no induction of innate immune responses. Thus, PTD-DRBD–mediated siRNA delivery allows efficient gene silencing in difficult-to-transfect primary cell types.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
de Fougerolles, A., Vornlocher, H.P., Maraganore, J. & Lieberman, J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat. Rev. Drug Discov. 6, 443–453 (2007).
Kim, D.H. & Rossi, J.J. Strategies for silencing human disease using RNA interference. Nat. Rev. Genet. 8, 173–184 (2007).
Bumcrot, D., Manoharan, M., Koteliansky, V. & Sah, D.W. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat. Chem. Biol. 2, 711–719 (2006).
Whitehead, K.A., Langer, R. & Anderson, D.G. Knocking down barriers: advances in siRNA delivery. Nat. Rev. Drug Discov. 8, 129–138 (2009).
Behlke, M.A. Chemical modification of siRNAs for in vivo use. Oligonucleotides 18, 305–320 (2008).
Gump, J.M. & Dowdy, S.F. TAT transduction: the molecular mechanism and therapeutic prospects. Trends Mol. Med. 13, 443–448 (2007).
Nakase, I. et al. Cellular uptake of arginine-rich peptides: roles for macropinocytosis and actin rearrangement. Mol. Ther. 10, 1011–1022 (2004).
Wadia, J.S., Stan, R.V. & Dowdy, S.F. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat. Med. 10, 310–315 (2004).
Turner, J.J. et al. RNA targeting with peptide conjugates of oligonucleotides, siRNA and PNA. Blood Cells Mol. Dis. 38, 1–7 (2007).
Meade, B.R. & Dowdy, S.F. Enhancing the cellular uptake of siRNA duplexes following noncovalent packaging with protein transduction domain peptides. Adv. Drug Deliv. Rev. 60, 530–536 (2008).
Bevilacqua, P.C. & Cech, T.R. Minor-groove recognition of double-stranded RNA by the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR. Biochemistry 35, 9983–9994 (1996).
Tian, B., Bevilacqua, P.C., Diegelman-Parente, A. & Mathews, M.B. The doublestranded-RNA-binding motif: interference and much more. Nat. Rev. Mol. Cell Biol. 5, 1013–1023 (2004).
Ryter, J.M. & Schultz, S.C. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 17, 7505–7513 (1998).
Boyer, L.A. et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956 (2005).
Henderson, J.K. et al. Preimplantation human embryos and embryonic stem cells show comparable expression of stage-specific embryonic antigens. Stem Cells 20, 329–337 (2002).
Hay, D.C., Sutherland, L., Clark, J. & Burdon, T. Oct-4 knockdown induces similar patterns of endoderm and trophoblast differentiation markers in human and mouse embryonic stem cells. Stem Cells 22, 225–235 (2004).
Zhang, W. et al. Inhibition of respiratory syncytial virus infection with intranasal siRNA nanoparticles targeting the viral NS1 gene. Nat. Med. 11, 56–62 (2005).
Judge, A.D. et al. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol. Ther. 13, 494–505 (2006).
Safran, M. et al. Mouse reporter strain for noninvasive bioluminescent imaging of cells that have undergone Cre-mediated recombination. Mol. Imaging 2, 297–302 (2003).
Miyoshi, H. et al. Development of a self-inactivating lentivirus vector. J. Virol. 72, 8150–8157 (1998).
Acknowledgements
We thank V. Nizet (UCSD) for PBMCs. The hES cell line HUES9 was kindly provided by D. Melton (HHMI, Harvard University). A.E. was funded by a Japan Society for the Promotion of Science Research Fellowships for Young Scientists. This work was supported by a Specialized Center of Research grant from the Leukemia & Lymphoma Society (S.F.D.), the Elsa U. Pardee Foundation (S.F.D.), the Howard Hughes Medical Institute (S.F.D.) and the California Institute for Regenerative Medicine (S.F.D.).
Author information
Authors and Affiliations
Contributions
A.E. and B.R.M. designed, purified PTD-DRBD and performed RNAi experiments. Y.-C.C performed PBMC experiments. C.T.F. performed hES cell culture. K.W. supervised hES cell culture. N.P. provided siRNAs reagents. S.F.D. supervised and analyzed data. A.E. and S.F.D. contributed to writing the manuscript, and all authors discussed and refined the manuscript.
Corresponding author
Ethics declarations
Competing interests
A.E., B.R.M. and S.F.D. are co-inventors on a patent application related to the method described in this publication, which has been licensed to Traversa Therapeutics. S.F.D. is the scientific founder of Traversa Therapeutics.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1 and 2 (PDF 758 kb)
Rights and permissions
About this article
Cite this article
Eguchi, A., Meade, B., Chang, YC. et al. Efficient siRNA delivery into primary cells by a peptide transduction domain–dsRNA binding domain fusion protein. Nat Biotechnol 27, 567–571 (2009). https://doi.org/10.1038/nbt.1541
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.1541
This article is cited by
-
Cytoplasmic delivery of siRNA using human-derived membrane penetration-enhancing peptide
Journal of Nanobiotechnology (2022)
-
CHIP ameliorates neuronal damage in H2O2-induced oxidative stress in HT22 cells and gerbil ischemia
Scientific Reports (2022)
-
Use of cell cultures in vitro to assess the uptake of long dsRNA in plant cells
In Vitro Cellular & Developmental Biology - Plant (2022)
-
Advances in oligonucleotide drug delivery
Nature Reviews Drug Discovery (2020)
-
Intranasal Drug Delivery into Mouse Nasal Mucosa and Brain Utilizing Arginine-Rich Cell-Penetrating Peptide-Mediated Protein Transduction
International Journal of Peptide Research and Therapeutics (2020)