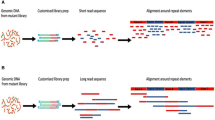
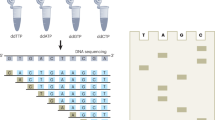
Abstract
The increasing availability of high-quality reference genomic sequences has created a demand for ways to survey the sequence differences present in individual genomes. Here we describe a DNA sequencing method based on hybridization of a universal panel of tiling probes. Millions of shotgun fragments are amplified in situ and subjected to sequential hybridization with short fluorescent probes. Long fragments of 200 bp facilitate unique placement even in large genomes. The sequencing chemistry is simple, enzyme-free and consumes only dilute solutions of the probes, resulting in reduced sequencing cost and substantially increased speed. A prototype instrument based on commonly available equipment was used to resequence the Bacteriophage λ and Escherichia coli genomes to better than 99.93% accuracy with a raw throughput of 320 Mbp/day, albeit with a significant number of small gaps attributed to losses in sample preparation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Sanger, F., Nicklen, S. & Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74, 5463–5467 (1977).
Prober, J.M. et al. A system for rapid DNA sequencing with fluorescent chain-terminating dideoxynucleotides. Science 238, 336–341 (1987).
Luckey, J.A. et al. High speed DNA sequencing by capillary electrophoresis. Nucleic Acids Res. 18, 4417–4421 (1990).
Venter, J.C. et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304, 66–74 (2004).
The International HapMap Consortium. A haplotype map of the human genome. Nature 437, 1299–1320 (2005).
Klein, R.J. et al. Complement factor H polymorphism in age-related macular degeneration. Science 308, 385–389 (2005).
Maraganore, D.M. et al. High-resolution whole-genome association study of Parkinson disease. Am. J. Hum. Genet. 77, 685–693 (2005).
Margulies, M. et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380 (2005).
Shendure, J. et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science 309, 1728–1732 (2005).
Blazej, R.G., Kumaresan, P. & Mathies, R.A. Microfabricated bioprocessor for integrated nanoliter-scale Sanger DNA sequencing. Proc. Natl. Acad. Sci. USA 103, 7240–7245 (2006).
Bennett, S.T., Barnes, C., Cox, A., Davies, L. & Brown, C. Toward the 1,000 dollars human genome. Pharmacogenomics 6, 373–382 (2005).
Shendure, J., Mitra, R.D., Varma, C. & Church, G.M. Advanced sequencing technologies: methods and goals. Nat. Rev. Genet. 5, 335–344 (2004).
Brenner, S. et al. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 18, 630–634 (2000).
Ghadessy, F.J., Ong, J.L. & Holliger, P. Directed evolution of polymerase function by compartmentalized self-replication. Proc. Natl. Acad. Sci. USA 98, 4552–4557 (2001).
Mitra, R.D. & Church, G.M. In situ localized amplification and contact replication of many individual DNA molecules. Nucleic Acids Res. 27, e34 (1999).
Bing, D.H. et al. Bridge amplification: a solid phase PCR system for the amplification and detection of allelic differences in single copy genes. Genetic Identity Conference Proceedings, Seventh International Symposium on Human Identification, Scottsdale, AZ, September 18–20, 1996 (Promega Corp., Madison, WI, 1996).
Braslavsky, I., Hebert, B., Kartalov, E. & Quake, S.R. Sequence information can be obtained from single DNA molecules. Proc. Natl. Acad. Sci. USA 100, 3960–3964 (2003).
Hyman, E.D. A new method of sequencing DNA. Anal. Biochem. 174, 423–436 (1988).
Ronaghi, M., Karamohamed, S., Pettersson, B., Uhlen, M. & Nyren, P. Real-time DNA sequencing using detection of pyrophosphate release. Anal. Biochem. 242, 84–89 (1996).
Metzker, M.L. et al. Termination of DNA synthesis by novel 3′-modified-deoxyribonucleoside 5′-triphosphates. Nucleic Acids Res. 22, 4259–4267 (1994).
Canard, B. & Sarfati, R.S. DNA polymerase fluorescent substrates with reversible 3′-tags. Gene 148, 1–6 (1994).
Hinds, D.A. et al. Whole-genome patterns of common DNA variation in three human populations. Science 307, 1072–1079 (2005).
Drmanac, R., Petrovic, N., Glisin, V. & Crkvenjakov, R. Sequencing of megabase plus DNA by hybridization: theory of the method. Genomics 4, 114–128 (1989).
Drmanac, S. et al. Accurate sequencing by hybridization for DNA diagnostics and individual genomics. Nat. Biotechnol. 16, 54–58 (1998).
Bains, W. & Smith, G.C. A novel method for nucleic acid sequence determination. J. Theor. Biol. 135, 303–307 (1988).
Lysov, Y.P., Florent'ev, V.L., Khorlin, A.A., Khrapko, K.R. & Shik, V.V. Determination of the nucleotide sequence of DNA using hybridization with oligonucleotides. A new method. Dokl. Akad. Nauk SSSR 303, 1508–1511 (1988).
Lizardi, P.M. et al. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat. Genet. 19, 225–232 (1998).
Koshkin, A.A. et al. LNA (Locked Nucleic Acids): synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron 54, 3607–3630 (1998).
Donachie, W.D. The cell cycle of Escherichia coli. Annu. Rev. Microbiol. 47, 199–230 (1993).
Ewing, B. & Green, P. Base-calling of automated sequencer traces using phred. ii. Error probabilities. Genome Res. 8, 186–194 (1998).
Shamir, R. & Tsur, D. Large scale sequencing by hybridization. J. Comput. Biol. 9, 413–428 (2002).
Drmanac, R. et al. Sequencing by hybridization (SBH): advantages, achievements, and opportunities. Adv. Biochem. Eng. Biotechnol. 77, 75–101 (2002).
Arratia, R., Martin, D., Reinert, G. & Waterman, M.S. Poisson process approximation for sequence repeats, and sequencing by hybridization. J. Comput. Biol. 3, 425–463 (1996).
Pe'er, I., Arbili, N. & Shamir, R. A computational method for resequencing long DNA targets by universal oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 99, 15492–15496 (2002).
Whiteford, N. et al. An analysis of the feasibility of short read sequencing. Nucleic Acids Res. 33, e171 (2005).
Lander, E.S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Church, G., Shendure, J. & Porreca, G. Sequencing thoroughbreds. Nat. Biotechnol. 24, 139 (2006).
Williams, R. et al. Amplification of complex gene libraries by emulsion PCR. Nat. Methods 3, 545–550 (2006).
Acknowledgements
We thank M. Nilsson and U. Landegren for early advice on RCA; J. Sagemark for initial bioinformatics analysis of the feasibility of the concept; P. Bérubé for the E. coli DNA preparation; M. Belouchi, P. van Eerdewegh, J. Hooper, B. Houle, R. Paulussen for helpful discussions; and P. Ernfors for advice and discussions. This work was supported by Swedish Research Council grant 522-2006-6511.
Author information
Authors and Affiliations
Contributions
A.P. developed the short LNA probes, participated in the development of sample preparation, the rolling-circle arrays and the hybridization cycle. G.B. participated in the development of sample preparation and the hybridization cycle. A.M. participated in the development of sample preparation and the rolling-circle arrays. P.L. participated in the development of image analysis and basecalling software and algorithms. E.H. developed the custom Peltier assembly, built the instrument and participated in the development of instrument control and image analysis software. S.L. conceived of the concept of shotgun SBH, participated in probe design, the development of sample preparation, rolling-circle arrays, hybridization cycle, and of the instrument control, image analysis and basecalling software; analyzed the experiments, directed the research and drafted the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors are former employees of Genizon Svenska AB, an affiliate of Genizon Biosciences Inc. (Montreal), which has also funded the research. The authors may under certain circumstances stand to receive royalty payments or similar benefits related to the technology presented in the paper.
Supplementary information
Supplementary Text and Figures
Supplementary Figs. 1–3, Tables 1–3 (PDF 478 kb)
Supplementary Data
Supplementary Data (XLS 527 kb)
Rights and permissions
About this article
Cite this article
Pihlak, A., Baurén, G., Hersoug, E. et al. Rapid genome sequencing with short universal tiling probes. Nat Biotechnol 26, 676–684 (2008). https://doi.org/10.1038/nbt1405
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt1405
This article is cited by
-
An Algorithm for Sequencing by Hybridization Based on an Alternating DNA Chip
Interdisciplinary Sciences: Computational Life Sciences (2018)
-
Single-molecule mechanical identification and sequencing
Nature Methods (2012)
-
Estimating accuracy of RNA-Seq and microarrays with proteomics
BMC Genomics (2009)
-
The challenges of sequencing by synthesis
Nature Biotechnology (2009)
-
High quality draft sequences for prokaryotic genomes using a mix of new sequencing technologies
BMC Genomics (2008)