Abstract
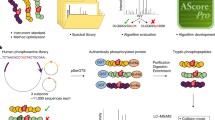
Data analysis and interpretation remain major logistical challenges when attempting to identify large numbers of protein phosphorylation sites by nanoscale reverse-phase liquid chromatography/tandem mass spectrometry (LC-MS/MS) (Supplementary Figure 1 online). In this report we address challenges that are often only addressable by laborious manual validation, including data set error, data set sensitivity and phosphorylation site localization. We provide a large-scale phosphorylation data set with a measured error rate as determined by the target-decoy approach, we demonstrate an approach to maximize data set sensitivity by efficiently distracting incorrect peptide spectral matches (PSMs), and we present a probability-based score, the Ascore, that measures the probability of correct phosphorylation site localization based on the presence and intensity of site-determining ions in MS/MS spectra. We applied our methods in a fully automated fashion to nocodazole-arrested HeLa cell lysate where we identified 1,761 nonredundant phosphorylation sites from 491 proteins with a peptide false-positive rate of 1.3%.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Kim, J.E., Tannenbaum, S.R. & White, F.M. Global phosphoproteome of HT-29 human colon adenocarcinoma cells. J. Proteome Res. 4, 1339–1346 (2005).
Cantin, G.T., Venable, J.D., Cociorva, D. & Yates, J.R., III . Quantitative phosphoproteomic analysis of the tumor necrosis factor pathway. J. Proteome Res. 5, 127–134 (2006).
Ballif, B.A., Villen, J., Beausoleil, S.A., Schwartz, D. & Gygi, S.P. Phosphoproteomic analysis of the developing mouse brain. Mol. Cell. Proteomics 3, 1093–1101 (2004).
Beausoleil, S.A. et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. USA 101, 12130–12135 (2004).
Ficarro, S.B. et al. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 20, 301–305 (2002).
Gruhler, A. et al. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell. Proteomics 4, 310–327 (2005).
Nuhse, T.S., Stensballe, A., Jensen, O.N. & Peck, S.C. Large-scale analysis of in vivo phosphorylated membrane proteins by immobilized metal ion affinity chromatography and mass spectrometry. Mol. Cell. Proteomics 2, 1234–1243 (2003).
Rush, J. et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 23, 94–101 (2005).
Collins, M.O. et al. Proteomic analysis of in vivo phosphorylated synaptic proteins. J. Biol. Chem. 280, 5972–5982 (2005).
Trinidad, J.C., Specht, C.G., Thalhammer, A., Schoepfer, R. & Burlingame, A.L. Comprehensive identification of phosphorylation sites in postsynaptic density preparations. Mol. Cell Proteomics 5, 914–922 (2006).
MacCoss, M.J. Computational analysis of shotgun proteomics data. Curr. Opin. Chem. Biol. 9, 88–94 (2005).
Peng, J., Elias, J.E., Thoreen, C.C., Licklider, L.J. & Gygi, S.P. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2, 43–50 (2003).
Elias, J.E., Gibbons, F.D., King, O.D., Roth, F.P. & Gygi, S.P. Intensity-based protein identification by machine learning from a library of tandem mass spectra. Nat. Biotechnol. 22, 214–219 (2004).
DeGnore, J.P. & Qin, J. Fragmentation of phosphopeptides in an ion trap mass spectrometer. J. Am. Soc. Mass Spectrom. 9, 1175–1188 (1998).
Schwartz, D. & Gygi, S.P. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 23, 1391–1398 (2005).
Elias, J.E., Haas, W., Faherty, B.K. & Gygi, S.P. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat. Methods 2, 667–675 (2005).
Pawson, T. & Scott, J.D. Protein phosphorylation in signaling—50 years and counting. Trends Biochem. Sci. 30, 286–290 (2005).
Ballif, B.A. et al. Quantitative phosphorylation profiling of the ERK/p90 ribosomal S6 kinase-signaling cassette and its targets, the tuberous sclerosis tumor suppressors. Proc. Natl. Acad. Sci. USA 102, 667–672 (2005).
Stemmann, O., Zou, H., Gerber, S.A., Gygi, S.P. & Kirschner, M.W. Dual inhibition of sister chromatid separation at metaphase. Cell 107, 715–726 (2001).
Syka, J.E. et al. Novel linear quadrupole ion trap/FT mass spectrometer: performance characterization and use in the comparative analysis of histone H3 post-translational modifications. J. Proteome Res. 3, 621–626 (2004).
Haas, W. et al. Optimization and use of peptide mass measurement accuracy in shotgun proteomics. Mol. Cell. Proteomics (in the press) (2006).
Acknowledgements
We thank David Chiang, James Candlin and the software developers at Sage-N-Research for early access to Sequest-Sorcerer and on-the-fly peptide reversal within the Sequest algorithm. We thank P. Everley, C. Bakalarski and B. Faherty for in-house software development and D. Schwartz for motif analysis. HeLa cell lysate was generously provided by M. Rape. This work was supported in part by grants from the National Institutes of Health (HG03456 and GM67945).
Author information
Authors and Affiliations
Contributions
S.A.B. conducted all experiments, carried out algorithm development and implementation, and data analysis. J.V. and S.A.G. provided analytical expertise for SCX chromatography and mass spectrometry. J.R. provided synthetic peptide libraries and immunoprecipitation data. S.P.G. provided overall experimental design and support.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
General Strategy for the large-scale analysis of protein phosphorylation. (PDF 757 kb)
Supplementary Fig. 2
Determining an effective search space using the target/decoy strategy. (PDF 863 kb)
Supplementary Fig. 3
Residue composition from data sets of known phosphorylation sites. (PDF 1768 kb)
Supplementary Fig. 4
Comparison of the Ascore versus Sequest scoring criteria for site localization. (PDF 3070 kb)
Supplementary Fig. 5
Comparison of the Ascore versus Mascot scoring criteria for site localization. (PDF 3256 kb)
Supplementary Fig. 6
Sequence logos (http://weblogo.berkeley.edu/) of the motifs identified in this large-scale data set shown in Supplementary table 1 with an Ascore > 15. (PDF 978 kb)
Supplementary Fig. 7
Biological processes of 362 of the 491 proteins identified in this experiment. (PDF 622 kb)
Supplementary Fig. 8
SDS-PAGE gel used in this experiment. (PDF 5115 kb)
Supplementary Table 1
Filtering criteria for the entire experiment. (PDF 1053 kb)
Supplementary Table 2
2,836 identified phosphopeptides from the entire experiment. (XLS 1649 kb)
Supplementary Table 3
Synthetic peptide libraries. (XLS 815 kb)
Supplementary Table 4
Immunoprecipitation experiments. (XLS 495 kb)
Rights and permissions
About this article
Cite this article
Beausoleil, S., Villén, J., Gerber, S. et al. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol 24, 1285–1292 (2006). https://doi.org/10.1038/nbt1240
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt1240
This article is cited by
-
Multi-omics analysis identifies drivers of protein phosphorylation
Genome Biology (2023)
-
ORC1 binds to cis-transcribed RNAs for efficient activation of replication origins
Nature Communications (2023)
-
DeepFLR facilitates false localization rate control in phosphoproteomics
Nature Communications (2023)
-
Mzion enables deep and precise identification of peptides in data-dependent acquisition proteomics
Scientific Reports (2023)
-
Glycopeptide database search and de novo sequencing with PEAKS GlycanFinder enable highly sensitive glycoproteomics
Nature Communications (2023)