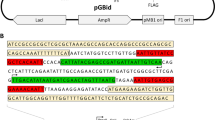
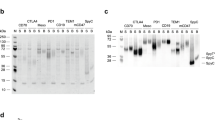
Abstract
We show here that the number of single-chain antibody fragments (scFv) presented on filamentous phage particles generated with antibody display phagemids can be increased by more than two orders of magnitude by using a newly developed helper phage (hyperphage). Hyperphage have a wild-type pIII phenotype and are therefore able to infect F+ Escherichia coli cells with high efficiency; however, their lack of a functional pIII gene means that the phagemid-encoded pIII–antibody fusion is the sole source of pIII in phage assembly. This results in an considerable increase in the fraction of phage particles carrying an antibody fragment on their surface. Antigen-binding activity was increased about 400-fold by enforced oligovalent antibody display on every phage particle. When used for packaging a universal human scFv library, hyperphage improved the specific enrichment factor obtained when panning on tetanus toxin. After two panning rounds, more than 50% of the phage were found to bind to the antigen, compared to 3% when conventional M13KO7 helper phage was used. Thus, hyperphage is particularly useful in stoichiometric situations, when there is little chance that a single phage will locate the desired antigen.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Hoogenboom, H.R. et al. Multi-subunit proteins on the surface of filamentous phage: methodologies for displaying antibody (Fab) heavy and light chains. Nucleic Acids Res. 19, 4133–4137 (1991).
Breitling, F. et al. A surface expression vector antibody screening. Gene 104, 147–153 (1991).
Marks, J.D. et al. By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J. Mol. Biol. 222, 581–597 (1991).
Barbas, C.F. et al. Assembly of combinatorial antibody libraries on phage surfaces: the gene III site. Proc. Natl. Acad. Sci. USA 88, 7978–7982 (1991).
Gavilondo, J.V. & Larrick, J.W. Antibody engineering at the millennium. BioTechniques 29, 128–145 (2000).
Fisch, I. et al. A strategy of exon shuffling for making large peptide repertoires displayed on filamentous bacteriophage. Proc. Natl. Acad. Sci. USA 93, 7761–7766 (1996).
Knappik, A. et al. Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CDRs randomized with trinucleotides. J. Mol. Biol. 296, 57–86 (2000).
Sblattero, D. & Bradburry, A. Exploiting recombination in single bacteria to make large phage antibody libraries. Nat. Biotechnol. 18, 75–80 (2000).
Griffiths, A.D. et al. Human anti-self antibodies with high specificity from phage display libraries. EMBO J. 12, 725–734 (1993)
Duenas, M. & Borrebaeck, C.A. Novel helper phage design: intergenic region affects the assembly of bacteriophages and the size of antibody libraries. FEMS Microbiol Lett. 125, 317–321 (1995)
Rakonjac, J., Jovanovic, G. & Model, P. Filamentous phage infection-mediated gene expression: construction and propagation of the gIII deletion mutant helper phage R408d3. Gene 198, 99–103 (1997)
Nelson, F.K., Friedman, S.M. & Smith, G.P. Filamentous phage DNA cloning vectors: a noninfective mutant with a nonpolar deletion in gene III. Virology 108, 338–350 (1981)
Crissman, J.W. & Smith, G.P. Gene-III protein of filamentous phages: evidence for a carboxyl-terminal domain with a role in morphogenesis. Virology 132, 445–455 (1984)
Dotto, G.P. & Zinder, N.D. Reduction of the minimal sequence for initiation of DNA synthesis by qualitative or quantitative changes of an initiator protein. Nature 311, 279–280 (1984).
Davis, N.G., Boeke, J.D. & Model, P. Fine structure of a membrane anchor domain. J. Mol. Biol. 181, 111–121 (1985).
Rondot, S. Präsentation von Peptidepitopen auf rekombinanten F-Pili für eine antigenspezifische Infektion von Bakterien (Inaugural-Dissertation). (University of Heidelberg, Germany; 1997).
Dübel, S. et al. A family of vectors for surface display and production of antibodies. Gene 128, 97–101 (1993).
Diederich, L., Rasmussen, L.J. & Messer, W. New cloning vectors for integration into the λ attachment site attB of the Escherichia coli chromosome. Plasmid 28, 14–24 (1992)
Welschof, M. et al. The antigen-binding domain of a human IgG-anti-F(ab′)2 autoantibody. Proc. Natl. Acad. Sci. USA 94, 1902–1907 (1997).
Micheel, B. et al. Production of monoclonal antibodies against epitopes of the main coat protein of filamentous fd phages. J. Immunol. Methods 171, 103–109 (1994).
Koch, J. & Dübel., S. Generation of antibody libraries from human donors. In Antibody engineering, (eds Kontermann, R. & Dübel, S.) (Springer Verlag, Heidelberg/New York, in press).
Yuan Q. et al. Molecular cloning, expression, and characterization of a functional single-chain Fv antibody to the mycotoxin zearalenone. Appl. Environ. Microbiol. 63, 263–269 (1997).
Schier, R. et al. Isolation of high-affinity monomeric human anti-c-erbB-2 single chain Fv using affinity-driven selection. J. Mol. Biol. 255, 28–43 (1996).
Smith, G.P. Filamentous phages as cloning vectors. Biotechnology 10, 61–83 (1988).
Kramer, A. & Schneider-Mergener, J. Synthesis of peptide libraries on continuous cellulose membranes. Methods Mol. Biol. 87, 25–39 (1997).
Sambrook, J., Fritsch, E.F. & Maniatis, T. Molecular cloning. A laboratory manual, Edn. 2. (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY; 1989).
Acknowledgements
The financial support from the Heidelberger Akademie der Wissenschaften (Germany) is gratefully acknowledged. We wish to thank M. Little for support, M. Bechtel from Genzyme Virotech GmbH for supplying the tetanus toxin, and Olaf Broders for helping with the experimental work. We are grateful for E.K.F. Bautz for his continuous support and suggestions.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rondot, S., Koch, J., Breitling, F. et al. A helper phage to improve single-chain antibody presentation in phage display. Nat Biotechnol 19, 75–78 (2001). https://doi.org/10.1038/83567
Received:
Revised:
Issue Date:
DOI: https://doi.org/10.1038/83567
This article is cited by
-
Construction and characterization of a novel miniaturized filamentous phagemid for targeted mammalian gene transfer
Microbial Cell Factories (2023)
-
M13 phage: a versatile building block for a highly specific analysis platform
Analytical and Bioanalytical Chemistry (2023)
-
Potential of Phage Display Antibody Technology for Cardiovascular Disease Immunotherapy
Journal of Cardiovascular Translational Research (2022)
-
Isolation and light chain shuffling of a Plasmodium falciparum AMA1-specific human monoclonal antibody with growth inhibitory activity
Malaria Journal (2021)