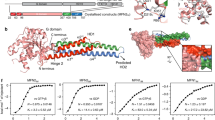
Abstract
Mitochondria are dynamic organelles that exchange contents and undergo remodelling during cyclic fusion and fission. Genetic mutations in MFN2 (the gene encoding mitofusin 2) interrupt mitochondrial fusion and cause the untreatable neurodegenerative condition Charcot-Marie-Tooth disease type 2A (CMT2A). It has not yet been possible to directly modulate mitochondrial fusion, in part because the structural basis of mitofusin function is not completely understood. Here we show that mitofusins adopt either a fusion-constrained or a fusion-permissive molecular conformation, directed by specific intramolecular binding interactions, and demonstrate that mitofusin-dependent mitochondrial fusion can be regulated in mouse cells by targeting these conformational transitions. On the basis of this model, we engineered a cell-permeant minipeptide to destabilize the fusion-constrained conformation of mitofusin and promote the fusion-permissive conformation, reversing mitochondrial abnormalities in cultured fibroblasts and neurons that harbour CMT2A-associated genetic defects. The relationship between the conformational plasticity of mitofusin 2 and mitochondrial dynamism reveals a central mechanism that regulates mitochondrial fusion, the manipulation of which can correct mitochondrial pathology triggered by defective or imbalanced mitochondrial dynamics.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Chan, D. C. Fusion and fission: interlinked processes critical for mitochondrial health. Annu. Rev. Genet. 46, 265–287 (2012)
Chen, H. et al. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 160, 189–200 (2003)
Kasahara, A., Cipolat, S., Chen, Y., Dorn, G. W., II & Scorrano, L. Mitochondrial fusion directs cardiomyocyte differentiation via calcineurin and Notch signaling. Science 342, 734–737 (2013)
Chen, H., McCaffery, J. M. & Chan, D. C. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell 130, 548–562 (2007)
Chen, H. et al. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell 141, 280–289 (2010)
Song, M., Mihara, K., Chen, Y., Scorrano, L. & Dorn, G. W., II . Mitochondrial fission and fusion factors reciprocally orchestrate mitophagic culling in mouse hearts and cultured fibroblasts. Cell Metab. 21, 273–285 (2015)
Bombelli, F. et al. Charcot-Marie-Tooth disease type 2A: from typical to rare phenotypic and genotypic features. JAMA Neurol. 71, 1036–1042 (2014)
Koshiba, T. et al. Structural basis of mitochondrial tethering by mitofusin complexes. Science 305, 858–862 (2004)
Low, H. H. & Löwe, J. A bacterial dynamin-like protein. Nature 444, 766–769 (2006)
Huang, P., Galloway, C. A. & Yoon, Y. Control of mitochondrial morphology through differential interactions of mitochondrial fusion and fission proteins. PLoS One 6, e20655 (2011)
Chen, H. et al. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 160, 189–200 (2003)
Chen, Y. & Dorn, G. W., II . PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science 340, 471–475 (2013)
Gong, G. et al. Parkin-mediated mitophagy directs perinatal cardiac metabolic maturation in mice. Science 350, aad2459 (2015)
Chen, H., Chomyn, A. & Chan, D. C. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J. Biol. Chem. 280, 26185–26192, (2005)
Detmer, S. A. & Chan, D. C. Complementation between mouse Mfn1 and Mfn2 protects mitochondrial fusion defects caused by CMT2A disease mutations. J. Cell Biol. 176, 405–414 (2007)
Calvo, J. et al. Genotype-phenotype correlations in Charcot-Marie-Tooth disease type 2 caused by mitofusin 2 mutations. Arch. Neurol. 66, 1511–1516 (2009)
Detmer, S. A., Vande Velde, C., Cleveland, D. W. & Chan, D. C. Hindlimb gait defects due to motor axon loss and reduced distal muscles in a transgenic mouse model of Charcot-Marie-Tooth type 2A. Hum. Mol. Genet. 17, 367–375 (2008)
Picard, M. et al. Trans-mitochondrial coordination of cristae at regulated membrane junctions. Nat. Commun. 6, 6259 (2015)
Sobieski, C., Jiang, X., Crawford, D. C. & Mennerick, S. Loss of local astrocyte support disrupts action potential propagation and glutamate release synchrony from unmyelinated hippocampal axon terminals in vitro. J. Neurosci. 35, 11105–11117 (2015)
Wang, W. et al. The ALS disease-associated mutant TDP-43 impairs mitochondrial dynamics and function in motor neurons. Hum. Mol. Genet. 22, 4706–4719 (2013)
Mochly-Rosen, D., Das, K. & Grimes, K. V. Protein kinase C, an elusive therapeutic target? Nat. Rev. Drug Discov. 11, 937–957 (2012)
Mackin, C. & Palacios, T. Large-scale sensor systems based on graphene electrolyte-gated field-effect transistors. Analyst 141, 2704–2711 (2016)
Qvit, N., Disatnik, M. H., Sho, E. & Mochly-Rosen, D. Selective phosphorylation inhibitor of delta protein kinase C–pyruvate dehydrogenase kinase protein–protein interactions: application for myocardial injury in vivo. J. Am. Chem. Soc. 138, 7626–7635 (2016)
Yang, J. et al. The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12, 7–8 (2015)
Acknowledgements
Supported by National Institutes of Health grants HL59888, HL128441 (G.W.D.); HL128071 (R.N.K. and G.W.D.); CA178394 (E.G.); MH078823 (S.M.); CA205262 (L.H.); and HL52141 (D.M.-R.); and American Cancer Society grant PF-15-135-01-CSM (S.K.D.). A.F. is supported by a fellowship from Societa' Italiana Ipertensione Arteriosa (SIIA). G.W.D. is the Philip and Sima K. Needleman-endowed Professor.
Author information
Authors and Affiliations
Contributions
G.W.D., D.M.-R., R.N.K., L.H., and S.M. conceived of or designed the research. G.W.D. wrote the manuscript. A.F., J.A.F., G.G. performed functional studies of minipeptides in MEFs. A.F., A.B., O.K. performed functional studies of minipeptides in neurons. A.F. and G.W.D. performed minipeptide mitochondrial colocalization. A.F. performed immunoblotting. N.Q. performed peptide-Mfn2 binding studies. O.K. performed Mfn2 GTPase assays. S.K.D., L.H., Y.C. performed Mfn2 FRET. E.G. and N.B. performed computational modelling and domain-interaction analyses. E.G., N.B., G.W.D., R.K. generated structural images.
Corresponding author
Ethics declarations
Competing interests
G.W.D. and D.M.-R. have applied for a patent for using fusion-modifying peptides in diseases with abnormal mitochondrial dynamics. D.M.-R. is a founder of MitoConix, but none of this work was supported by or performed in collaboration with the company. The other authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks R. Baloh, R. Lightowlers and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Figure 1 Interacting domains in human mitofusins.
a. Amino-acid alignment of human MFN1 and MFN2. Conservative amino-acid substitutions are indicated by plus symbols. MFN domains are colour-coded as in Fig. 1a: purple is the GTPase domain, red is HR1, black is transmembrane, green is HR2. Yellow amino acids in HR1 and red amino acids in HR2 are residues central to inter- or intramolecular binding. Areas where local unfolding of HR1 and HR2 α-helices occurs are highlighted yellow–green. b. Antiparallel intermolecular MFN2 HR2–HR2 binding as predicted in ref. 8. c. Intramolecular MFN2 HR1–HR2 binding as depicted in Fig. 1c and Supplementary Video 2.
Extended Data Figure 2 Adenovirally-expressed Gly-substituted mini-peptides derived from MFN2 HR1 specifically regulate Mfn-mediated mitochondrial fusion.
a, Changes in mitochondrial aspect ratio provoked by the different Mfn expression profiles of MP1Gly (adeno-367-384Gly) and MP2Gly (adeno-398-418Gly) in MEFs, compared to MEFs transfected with wild-type Mfn2 (ad-WT Mfn2) and MP-HR1 (adeno-HR1). Each point is the aspect ratio of an individual mitochondrion; group data are mean ± s.e.m.; *P < 0.05 versus MEFs transfected with empty adenovirus (adeno-null) by ANOVA. Exact numbers of mitochondria measured from four or five separate experiments per condition are shown. b, Mitochondrial polarization status and aspect ratio in wild-type and Mfn2-knockout MEFs expressing wild-type MFN2 or fragments thereof. Each point is the mean of approximately 20 mitochondria from approximately 5 cells in the number of independent experiments indicated on the graph. Group data are mean ± s.e.m.; *P < 0.05 versus MEF mitochondria transfected with empty adenovirus vector by ANOVA. Representative (of 15–40 images per group) merged confocal images of wild-type MEFs transfected with MFN2, or the two biologically active peptides are to the right. Scale bar, 10 μm. c. MitoTracker Green and mCherry Parkin (top) or Lyso-Red (bottom) co-stained wild-type or Mfn2-knockout MEFs before (left) or 1 h after (right) treatment with 50 nM FCCP. Group data are mean ± s.e.m.; there were no significant differences between groups (ANOVA; n = 3 independent experiments per condition). Scale bars, 10 μm.
Extended Data Figure 3 Effects of TAT-conjugated minipeptides on MEFs.
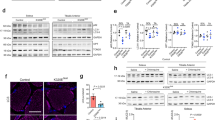
a, Merged confocal images (representative of 30) of mitochondria in MEFs 2 h after application of TAT–MP1Gly (367-384Gly; top) or TAT–MP2Gly (398-418Gly; bottom). Complete dose–response relationship is in Fig. 2d. b, Immunoblot analysis of mitochondrial dynamics proteins 4 h after application of TAT–MP1Gly (367-384) or TAT–MP2Gly (398-418). Opa1 is the inner mitochondrial membrane fusion protein Optic atrophy 1; Drp1 is the fission protein Dynamin-related protein 1. Opa1 immunoblot was co-probed for Mfn2. c, Immunoblot quantification. Data are mean ± s.e.m.; n = 4 for Mfn2; n = 2 for all other proteins. Uncropped original blots are in supplementary information Fig. 1.
Extended Data Figure 4 MFN2 HR1-derived TAT-minipeptides do not suppress GTPase activity, impair mitochondrial polarization, or increase cell death.
a. Relative GTPase activity of recombinant MFN2 is shown with respect to a no-Peptide control. Background signal from no GTP blank was subtracted from each value before normalization. Data are mean ± s.d. of three independent experiments. b, c, Mitochondrial depolarization was assessed as loss of TMRE staining (b) and cell death was assessed as ethidium bromide uptake (c) in wild-type MEFs cultured in 4,500 mg l−1 glucose (left) or galactose (right) treated with or without 1 μM TAT-mini-peptide for 24 h. n = 3 or 4 independent experiments per condition. All group data in panels a–c are mean ± s.e.m. There were no significant differences between TAT-mini-peptide treatment groups (ANOVA).
Extended Data Figure 5 TAT–MP1Gly rescues mitochondrial dysmorphology in a cell model of autosomal recessive CMT2A.
Representative (of 15–20 per group) confocal micrographs of Mfn2-knockout MEFs with stained mitochondria (green) and nuclei (blue) knockout (KO) at baseline or 24 h after application of mini-peptide (left); a representative normal MEF is shown for comparison. Mean normal MEF mitochondrial aspect ratio is 6.2. Group data are mean ± s.e.m.; *P < 0.05 versus control (ANOVA), n = 3 or 4 per group. Scale bar, 10 μm.
Extended Data Figure 6 Minipeptide effects on dysfunctional MFN2 mutants.
Wild-type MFN2 or the indicated mutants were expressed for 48 h using adenovirus vectors (ad) in Mfn-null MEFs (Mfn1/Mfn2 double knockout (DKO)). TAT-conjugated minipeptides or vehicle (sterile water) were added and mitochondrial morphometry assessed 24 h later. Images (representative of 15 per group) are of merged MitoTracker Green (green) and TMRE (red; stains active, polarized mitochondria) channels. Scale bars, 10 μm. Quantitative data for aspect ratio are shown below (n = 3 per condition). Group data are mean ± s.e.m.; *P < 0.05 versus same-group Ctrl (ANOVA).
Extended Data Figure 7 Wild-type MFN2 expression to parallel MFN2T105M expression studies.
a, Immunoblot analysis of mitochondrial dynamics proteins in cultured MEFs transduced with adenovirus-transfected βgal (viral control) or adenovirus-transfected human MFN2. MFN2 expression increased fivefold over uninfected Ctrl; apparent increased Mfn1 signal is cross immunoreactivity with MFN2 as shown by slightly faster migration. b, Merged confocal images (MitoTracker Green and TMRE; representative of 15 per group) of MEFs expressing wild-type MFN2 and treated with TAT-minipeptides as shown. Quantitative data (n = 3 independent experiments) are on the right. Group data are mean ± s.e.m.; *P < 0.05 versus control by one-way ANOVA. Scale bars, 10 μm. c, Immunoblot quantifications for panel a. d, Immunoblot quantifications for Fig. 4b, for comparison. n = 4 for MFN2; n = 2 for all other proteins. y axis scales are identical except MFN2 in c, which is expanded to accommodate high (5× normal) ad-WT MFN2 expression level.
Extended Data Figure 8 TAT–MP1Gly corrects mitochondrial fragmentation provoked by Mfn2K109A in cultured rat motor neurons.
Representative (of >20 per group) anti-TOM20/Hoechst images of formalin-fixed cultured neurons transfected with genes encoding Mfn2K109A (top) or wild-type Mfn2 (bottom) and treated for 24 h with 1 μM TAT control (Ctrl) or TAT–MP1Gly minipeptide. Scale bars, 10 μm. Experimental n number per treatment group is shown on the graph. Data are mean ± s.e.m. *P < 0.05 versus Ctrl (Student’s t-test).
Extended Data Figure 9 TAT–MP1Gly reverses mitochondrial pathology in cultured MFN2T105M mouse neurons.
Live-cell confocal imaging of MitoGFP (green), TMRE (red) and Hoechst (blue nuclei) stained mouse hippocampal neurons (representative of >30 images per group). Normal control neurons from nontransgenic (NTG) mouse pups are on top; MFN2T105M fl/st transgenic mouse neurons are on bottom. Adeno-MitoGFP was added at low titers to enhance visualization of individual neurons. Note mitochondrial fragmentation and clumping after Cre-induced induction of MFN2T105M, and reversal after 24 h of TAT–MP1Gly treatment. Scale bars, 20 μm.
Supplementary information
Supplementary Figure
This file contains immunoblot source data. (JPG 1145 kb)
Intermolecular human Mfn2 HR2-HR2 binding with full rotation
Purple is GTPase domain, red is HR1, black is transmembrane domain, green is HR2. (MOV 18284 kb)
Intramolecular human Mfn2 HR1-HR2 binding with full rotation
Purple is GTPase domain, red is HR1, black is transmembrane domain, green is HR2. (MPG 16986 kb)
Folded tethering non-permissive conformation of hMfn2 with full rotation.
Purple is GTPase domain, red is HR1, black is transmembrane domain, green is HR2. (MOV 56785 kb)
Unfolded, extended tethering permissive conformation of hMfn2 with full biaxial rotation.
Purple is GTPase domain, red is HR1, black is transmembrane domain, green is HR2. (MOV 23663 kb)
Rights and permissions
About this article
Cite this article
Franco, A., Kitsis, R., Fleischer, J. et al. Correcting mitochondrial fusion by manipulating mitofusin conformations. Nature 540, 74–79 (2016). https://doi.org/10.1038/nature20156
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature20156
This article is cited by
-
Targeting mitochondrial shape: at the heart of cardioprotection
Basic Research in Cardiology (2023)
-
Mitophagy for cardioprotection
Basic Research in Cardiology (2023)
-
Small molecule agonist of mitochondrial fusion repairs mitochondrial dysfunction
Nature Chemical Biology (2023)
-
Clinical and genetic features of a cohort of patients with MFN2-related neuropathy
Scientific Reports (2022)
-
Modulating mitofusins to control mitochondrial function and signaling
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.