Abstract
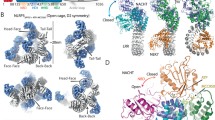
Inflammasomes are cytosolic innate immune complexes that activate caspase-1 following detection of pathogenic and endogenous dangers1,2,3,4,5, and NACHT-, leucine-rich repeat (LRR)- and pyrin domain (PYD)-containing protein 3 (NLRP3) is an inflammasome sensor of membrane damage highly important in regard to the induction of inflammation2,6,7. Here we report cryogenic electron microscopy structures of disc-shaped active NLRP3 oligomers in complex with adenosine 5′-O-(3-thio)triphosphate, the centrosomal NIMA-related kinase 7 (NEK7) and the adaptor protein ASC, which recruits caspase-1. In these NLRP3–NEK7–ASC complexes, the central NACHT domain of NLRP3 assumes an ATP-bound conformation in which two of its subdomains rotate by about 85° relative to the ADP-bound inactive conformation8,9,10,11,12. The fish-specific NACHT-associated domain conserved in NLRP3 but absent in most NLRPs13 becomes ordered in its key regions to stabilize the active NACHT conformation and mediate most interactions in the disc. Mutations on these interactions compromise NLRP3-mediated caspase-1 activation. The N-terminal PYDs from all NLRP3 subunits combine to form a PYD filament that recruits ASC PYD to elicit downstream signalling. Surprisingly, the C-terminal LRR domain and the LRR-bound NEK7 do not participate in disc interfaces. Together with previous structures of an inactive NLRP3 cage in which LRR–LRR interactions play an important role8,9,10,11, we propose that the role of NEK7 is to break the inactive cage to transform NLRP3 into the active NLRP3 inflammasome disc.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Atomic coordinates of the tenfold NLRP3–NEK7 assembly structure and NLRP3 PYD filament have been deposited in PDB under accession nos. 8EJ4 and 8ERT, respectively. Corresponding cryo-EM density maps have been deposited in the Electron Microscopy Data Bank under accession nos. EMD-28175 and EMD-28560, respectively. All other data are available from the corresponding authors on reasonable request.
References
Wang, L., Sharif, H., Vora, S. M., Zheng, Y. & Wu, H. Structures and functions of the inflammasome engine. J. Allergy Clin. Immunol. 147, 2021–2029 (2021).
Swanson, K. V., Deng, M. & Ting, J. P.-Y. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 19, 477–489 (2019).
Gaidt, M. M. & Hornung, V. The NLRP3 inflammasome renders cell death pro-inflammatory. J. Mol. Biol. 430, 133–141 (2018).
Nozaki, K., Li, L. & Miao, E. A. Innate sensors trigger regulated cell death to combat intracellular infection. Annu. Rev. Immunol. 40, 469–498 (2022).
Deets, K. A. & Vance, R. E. Inflammasomes and adaptive immune responses. Nat. Immunol. 22, 412–422 (2021).
Coll, R. C. et al. MCC950 directly targets the NLRP3 ATP-hydrolysis motif for inflammasome inhibition. Nat. Chem. Biol. 15, 556–559 (2019).
Tapia-Abellán, A. et al. MCC950 closes the active conformation of NLRP3 to an inactive state. Nat. Chem. Biol. 15, 560–564 (2019).
Sharif, H. et al. Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome. Nature 570, 338–343 (2019).
Andreeva, L. et al. NLRP3 cages revealed by full-length mouse NLRP3 structure control pathway activation. Cell 184, 6299–6312.e22 (2021).
Hochheiser, I. V. et al. Structure of the NLRP3 decamer bound to the cytokine release inhibitor CRID3. Nature 604, 184–189 (2022).
Ohto, U. et al. Structural basis for the oligomerization-mediated regulation of NLRP3 inflammasome activation. Proc. Natl Acad. Sci. USA 119, e2121353119 (2022).
Dekker, C. et al. Crystal structure of NLRP3 NACHT domain with an inhibitor defines mechanism of inflammasome inhibition. J. Mol. Biol. 433, 167309 (2021).
Tapia-Abellán, A. et al. Sensing low intracellular potassium by NLRP3 results in a stable open structure that promotes inflammasome activation. Sci. Adv. 7, eabf4468 (2021).
Muñoz-Planillo, R. et al. K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity 38, 1142–1153 (2013).
Magupalli, V. G. et al. HDAC6 mediates an aggresome-like mechanism for NLRP3 and pyrin inflammasome activation. Science 369, eaas8995 (2020).
Chen, J. & Chen, Z. J. PtdIns4P on dispersed trans-Golgi network mediates NLRP3 inflammasome activation. Nature 564, 71–76 (2018).
Li, X. et al. MARK4 regulates NLRP3 positioning and inflammasome activation through a microtubule-dependent mechanism. Nat. Commun. 8, 15986 (2017).
Lu, A. et al. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell 156, 1193–1206 (2014).
Hochheiser, I. V. et al. Directionality of PYD filament growth determined by the transition of NLRP3 nucleation seeds to ASC elongation. Sci. Adv. 8, eabn7583 (2022).
Li, Y. et al. Cryo-EM structures of ASC and NLRC4 CARD filaments reveal a unified mechanism of nucleation and activation of caspase-1. Proc. Natl Acad. Sci. USA 115, 10845–10852 (2018).
Lu, A. et al. Molecular basis of caspase-1 polymerization and its inhibition by a new capping mechanism. Nat. Struct. Mol. Biol. 23, 416–425 (2016).
He, Y., Zeng, M. Y., Yang, D., Motro, B. & Núñez, G. NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature 530, 354–357 (2016).
Schmid-Burgk, J. L. et al. A genome-wide CRISPR (clustered regularly interspaced short palindromic repeats) screen identifies NEK7 as an essential component of NLRP3 inflammasome activation. J. Biol. Chem. 291, 103–109 (2016).
Shi, H. et al. NLRP3 activation and mitosis are mutually exclusive events coordinated by NEK7, a new inflammasome component. Nat. Immunol. 17, 250–258 (2016).
Martinon, F., Burns, K. & Tschopp, J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol. Cell 10, 417–426 (2002).
Ding, J. et al. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 535, 111–116 (2016).
Liu, X. et al. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 535, 153–158 (2016).
He, W.-t et al. Gasdermin D is an executor of pyroptosis and required for interleukin-1β secretion. Cell Res. 25, 1285–1298 (2015).
Kayagaki, N. et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526, 666–671 (2015).
Shi, J. et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665 (2015).
Lieberman, J., Wu, H. & Kagan, J. C. Gasdermin D activity in inflammation and host defense. Sci. Immunol. 4, eaav1447 (2019).
Gaidt, M. M. & Hornung, V. Pore formation by GSDMD is the effector mechanism of pyroptosis. EMBO J. 35, 2167–2169 (2016).
Booshehri, L. M. & Hoffman, H. M. Caps and NLRP3. J. Clin. Immunol. 39, 277–286 (2019).
Touitou, I. et al. Infevers: an evolving mutation database for auto‐inflammatory syndromes. Hum. Mutat. 24, 194–198 (2004).
Mangan, M. S. et al. Targeting the NLRP3 inflammasome in inflammatory diseases. Nat. Rev. Drug Discov. 17, 588–606 (2018).
Duncan, J. A. et al. Cryopyrin/NALP3 binds ATP/dATP, is an ATPase, and requires ATP binding to mediate inflammatory signaling. Proc. Natl Acad. Sci. USA 104, 8041–8046 (2007).
Richards, M. W. et al. An autoinhibitory tyrosine motif in the cell-cycle-regulated Nek7 kinase is released through binding of Nek9. Mol. Cell 36, 560–570 (2009).
Zhang, L. et al. Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science 350, 404–409 (2015).
Hu, Z. et al. Structural and biochemical basis for induced self-propagation of NLRC4. Science 350, 399–404 (2015).
Hu, Z. et al. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science 341, 172–175 (2013).
Tenthorey, J. L. et al. The structural basis of flagellin detection by NAIP5: a strategy to limit pathogen immune evasion. Science 358, 888–893 (2017).
Yang, X. et al. Structural basis for specific flagellin recognition by the NLR protein NAIP5. Cell Res. 28, 35–47 (2018).
Sandall, C. F., Ziehr, B. K. & MacDonald, J. A. ATP-binding and hydrolysis in inflammasome activation. Molecules 25, 4572 (2020).
Zhou, M. et al. Atomic structure of the apoptosome: mechanism of cytochrome c-and dATP-mediated activation of Apaf-1. Genes Dev. 29, 2349–2361 (2015).
Yan, N. et al. Structure of the CED-4–CED-9 complex provides insights into programmed cell death in Caenorhabditis elegans. Nature 437, 831–837 (2005).
Qi, S. et al. Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4. Cell 141, 446–457 (2010).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Li, P. et al. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell 91, 479–489 (1997).
Schmacke, N. A. et al. IKKbeta primes inflammasome formation by recruiting NLRP3 to the trans-Golgi network. Immunity https://doi.org/10.1016/j.immuni.2022.10.021 (2022).
Hafner-Bratkovič, I. et al. NLRP3 lacking the leucine-rich repeat domain can be fully activated via the canonical inflammasome pathway. Nat. Commun. 9, 5182 (2018).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Shen, C. et al. Molecular mechanism for NLRP6 inflammasome assembly and activation. Proc. Natl Acad. Sci. USA 116, 2052–2057 (2019).
Tunyasuvunakool, K. et al. Highly accurate protein structure prediction for the human proteome. Nature 596, 590–596 (2021).
Goddard, T. D., Huang, C. C. & Ferrin, T. E. Visualizing density maps with UCSF Chimera. J. Struct. Biol. 157, 281–287 (2007).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
DeLano, W. L. Pymol: an open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 40, 82–92 (2002).
Acknowledgements
We thank H. Sharif, L. Andreeva, M. Mahammad, L. Wang and L. David for earlier work on this project, and Wu laboratory members for useful discussions. We thank Z. Yang for making lentiviruses and reconstitution of NLRP3 mutants into NLRP3–/– iBMDMs. For cryo-EM data collection we thank R. Walsh, S. Sterling, M. Megan, S. Rawson and Z. Li at the Harvard Cryo-EM Center for Structural Biology, and K. Song and J. Chang at the Cryo-EM Core Facility at University of Massachusetts Medical School. We thank K. Fitzgerald at University of Massachusetts Medical School for NLRP3–/– iBMDMs. We also thank R. Tomaino and Taplin Biological Mass Spectrometry Facility for protein analysis. Our research used software and computing support at SBGrid. This work was supported by US National Institutes of Health (nos. R01AI124491 to H.W. and R21AR079766 to V.G.M.), a postdoctoral fellowship from the KidneyCure Foundation (to L.X.) and a Faculty Career Development Fellowship from OFD/BTREC/CTREC Program of Boston Children’s Hospital (to V.G.M.).
Author information
Authors and Affiliations
Contributions
H.W. and L.X. conceived the study. V.G.M. generated and carried out initial expression of NLRP3 and NEK7 constructs by immunoblotting. L.X. designed and performed all experiments and H.W. supervised the project. L.X. and H.W. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
H.W. is a cofounder of Ventus Therapeutics. L.X. and V.G.M. declare no competing interests.
Peer review
Peer review information
Nature thanks Edward Miao and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
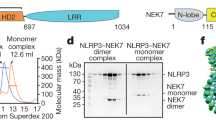
Extended Data Fig. 1 Preparation and characterization of the NLRP3-NEK7 complex.
a, Gel filtration profile of the NLRP3-NEK7 complex showing the two peaks. b, SDS-polyacrylamide gel electrophoresis (PAGE) of gel filtration fractions for the NLRP3-NEK7 complex. Locations of the two component proteins are labeled. c, Representative negative-staining images from the two NLRP3-NEK7 complex peaks. d, Gel filtration profile of the NLRP3-NEK7 complex with ATPγS showing one major peak. e, SDS-PAGE of the gel filtration fractions for the NLRP3-NEK7 complex with ATPγS. Locations of the two component proteins are labeled. f, A representative negative-staining image from the peak containing NLRP3, NEK7 and ATPγS. g, A representative cryo-EM image from the peak containing NLRP3, NEK7 and ATPγS. Experiments in (a-g) were repeated at least three times. h, 2D classes from cryo-EM images of the NLRP3-NEK7 complex with ATPγS. No full disk was observed.
Extended Data Fig. 2 Preparation and characterization of NLRP3-NEK7-ASC inflammasome complexes.
a, Gel filtration profile of the NLRP3-NEK7-ASC complex. b, SDS- PAGE of the near void peak from the gel filtration chromatography. Locations of the three component proteins are labelled. c, Mass spectrometry that detected ASC PYD in the band in (b). d, A representative cryo-EM image from a sample containing NLRP3, NEK7, ATPγS, and 1:1 ASC PYD. Experiments in (a-b) and (d) were repeated at least three times.
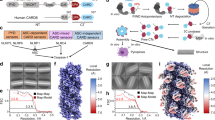
Extended Data Fig. 3 Flow chart for cryo-EM data processing of the NLRP3-NEK7-ASC inflammasome complex.
Data processing details can be found in Methods.
Extended Data Fig. 4 Local resolution distributions and Fourier shell correlation (FSC) curves.
a-c, The C10 NLRP3-NEK7-ASC complex (a), the C11 NLRP3-NEK7-ASC complex (b), and the partial NLRP3-NEK7 disk containing primarily 5 subunits (c).
Extended Data Fig. 5 PYD helical filament.
a-c, Side views of the cryo-EM maps of the C10 NLRP3-NEK7-ASC complex (a), the C11 NLRP3-NEK7-ASC complex (b) and the partial disk of the NLRP3-NEK7 complex (c). d, Local resolution distribution and Fourier shell correlation (FSC) curve of the NLRP3 PYD filament.
Extended Data Fig. 6 Close-up evaluation of the C10 NLRP3-NEK7-ASC complex map quality.
a-h, Cryo-EM maps in individual regions (labelled) superimposed with the final model.
Extended Data Fig. 7 Conformational change and comparison with NLRC4.
a, Superposition of active NLRP3 (in domain colours) and inactive NLRP3 (PDB: 6NPY in grey) by the FISNA-NBD-HD1 domain. The WHD domain of inactive NLRP3 situates behind the superimposed HD1, different from that of active NLRP3. b, Superposition of active NLRP3 and inactive NLRP3 (PDB: 6NPY in grey), showing the LRR domain and NEK7 of the inactive NLRP3-NEK7 complex would have been in clash with a neighbouring NLRP3 molecule. c, Superposition of active NLRP3 with active NLRC4 (PDB: 3JBL in yellow) by the FISNA-NBD-HD1 domain. d, Superposition of the FISNA domain from active NLRP3 (salmon), inactive NLRP3 (PDB: 6NPY in green), active NLRC4 (PDB: 3JBL in cyan), inactive NLRC4 (PDB: 4KXF in grey), and active NAIP5 (6B5B in yellow).
Extended Data Fig. 8 Mapping of the CAPS mutations on active NLRP3, and comparison of active NLRP3 and NLRC4.
a, Locations of CAPS mutation sites on the active NLRP3 structure. Two views are shown. b, A list of the mutations, their domain location and potential structural effects. c, Superposition of the WHD from active NLRP3 (magenta) and inactive NLRP3 (PDB: 6NPY in grey), showing the formation of a β-hairpin in the active state. d, Superposition of the WHD from active NLRP3 (magenta) and active NLRC4 (PDB: 3JBL in yellow), showing lack of the β-hairpin in NLRC4. e, Superposition of one NLRP3 subunit in two neighbouring NLRP3 subunits in a disk (coloured by domains) with one NLRC4 subunit in two neighbouring NLRC4 subunits in a disk (PDB: 3JBL in yellow). The second NLRC4 subunit needs to rotate by 31.7° to align with the second NLRP3 subunit.
Extended Data Fig. 9 A structure-derived model for potential NEK7-independent, and LRR-deleted NLRP3 inflammasome activation.
a, Under certain conditions such as those marked in the schematic (e.g. high NLRP3 expression level or certain effects from priming), monomeric NLRP3 or destabilized caged NLRP3 may directly form the active NLRP3 disk with ASC upon stimulation. b, For overexpressed LRR-deleted NLRP3, stimulation would likely directly induce the activating conformational change to allow the assembly of the inflammasome disk with ASC.
Supplementary information
Supplementary Information
Supplementary Fig. 1.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xiao, L., Magupalli, V.G. & Wu, H. Cryo-EM structures of the active NLRP3 inflammasome disc. Nature 613, 595–600 (2023). https://doi.org/10.1038/s41586-022-05570-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-05570-8
This article is cited by
-
Structural basis for the oligomerization-facilitated NLRP3 activation
Nature Communications (2024)
-
Mechanistic insights from inflammasome structures
Nature Reviews Immunology (2024)
-
Structural basis of the human NAIP/NLRC4 inflammasome assembly and pathogen sensing
Nature Structural & Molecular Biology (2024)
-
Drugging the NLRP3 inflammasome: from signalling mechanisms to therapeutic targets
Nature Reviews Drug Discovery (2024)
-
The NLRP3 inflammasome: a vital player in inflammation and mediating the anti-inflammatory effect of CBD
Inflammation Research (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.