Abstract
Parkinson’s disease (PD) is the most common movement disorder, with resting tremor, rigidity, bradykinesia and postural instability being major symptoms1. Neuropathologically, it is characterized by the presence of abundant filamentous inclusions of α-synuclein in the form of Lewy bodies and Lewy neurites in some brain cells, including dopaminergic nerve cells of the substantia nigra2. PD is increasingly recognised as a multisystem disorder, with cognitive decline being one of its most common non-motor symptoms. Many patients with PD develop dementia more than 10 years after diagnosis3. PD dementia (PDD) is clinically and neuropathologically similar to dementia with Lewy bodies (DLB), which is diagnosed when cognitive impairment precedes parkinsonian motor signs or begins within one year from their onset4. In PDD, cognitive impairment develops in the setting of well-established PD. Besides PD and DLB, multiple system atrophy (MSA) is the third major synucleinopathy5. It is characterized by the presence of abundant filamentous α-synuclein inclusions in brain cells, especially oligodendrocytes (Papp-Lantos bodies). We previously reported the electron cryo-microscopy structures of two types of α-synuclein filament extracted from the brains of individuals with MSA6. Each filament type is made of two different protofilaments. Here we report that the cryo-electron microscopy structures of α-synuclein filaments from the brains of individuals with PD, PDD and DLB are made of a single protofilament (Lewy fold) that is markedly different from the protofilaments of MSA. These findings establish the existence of distinct molecular conformers of assembled α-synuclein in neurodegenerative disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Berg, D. et al. Time to redefine Parkinson’s disease? Introductory statement of the MDS taskforce on the definition of Parkinson’s disease. Mov. Disord. 29, 454–462 (2014).
Goedert, M., Spillantini, M. G., Del Tredici, K. & Braak, H. 100 years of Lewy pathology. Nature Rev. Neurol. 9, 13–24 (2013).
Aarsland, D., Andersen, K., Larsen, J. P., Lolk, A. & Kragh-Sørensen, P. Prevalence and characteristics of dementia in Parkinson disease: an 8-year prospective study. Arch. Neurol. 60, 387–392 (2003).
McKeith, I. G. et al. Diagnosis and management of dementia with Lewy bodies. Fourth consensus report of the DLB consortium. Neurology 879, 88–100 (2017).
Fanciulli, A. & Wenning, G. K. Multiple system atrophy. N. Engl. J. Med. 372, 249–263 (2015).
Schweighauser, M. et al. Structures of α-synuclein filaments from multiple system atrophy. Nature 585, 464–469 (2020).
Polymeropoulos, M. H. et al. Mutation in the α-synuclein gene identified in families with Parkinson’s disease. Science 276, 2045–2047 (1997).
Singleton, A. B. et al. α-Synuclein locus triplication causes Parkinson’s disease. Science 302, 841 (2003).
Zarranz, J. J. et al. The new mutation, E46K, of alpha-synuclein causes Parkinson’s disease and Lewy body dementia. Ann. Neurol. 55, 164–173 (2004).
Nalls, M. A. et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nature Genet. 46, 989–993 (2014).
Davidson, W. S., Jonas, A., Clayton, D. F. & George, J. M. Stabilization of α-synuclein secondary structure upon binding to synthetic membranes. J. Biol. Chem. 273, 9443–9449 (1998).
Ueda, K. et al. Molecular cloning of cDNA encoding an unrecognized component of amyloid in Alzheimer disease. Proc. Natl Acad. Sci. USA 90, 11282–11286 (1993).
Li, H. T., Du, H. N., Tang, L., Hu, J. & Hu, H. Y. Structural transformation and aggregation of human α-synuclein in trifluoroethanol: non-amyloid component sequence is essential and β-sheet formation is prerequisite to aggregation. Biopolymers 64, 221–226 (2002).
Crowther, R. A., Jakes, R., Spillantini, M. G. & Goedert, M. Synthetic filaments assembled from C-terminally truncated α-synuclein. FEBS Lett. 436, 309–312 (1998).
Conway, K. A., Harper, J. D. & Lansbury, P. T. Fibrils formed in vitro from α-synuclein and two mutant forms linked to Parkinson’s disease are typical amyloid. Biochemistry 39, 2525–2563 (2000).
Serpell, L. C., Berriman, J., Jakes, R., Goedert, M. & Crowther, R. A. Fiber diffraction of synthetic α-synuclein filaments shows amyloid-like cross-β conformation. Proc. Natl Acad. Sci. USA 97, 4897–4902 (2000).
Miake, H., Mizusawa, H., Iwatsubo, T. & Hasegawa, M. Biochemical characterization of the core structure of α-synuclein filaments. J. Biol. Chem. 277, 19213–19219 (2002).
Mougenot, A. L. et al. Prion-like acceleration of a synucleinopathy in a transgenic mouse model. Neurobiol. Aging 33, 2225–2228 (2012).
Luk, K. C. et al. Pathological α-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science 338, 949–953 (2012).
Masuda-Suzukake, M. et al. Prion-like spreading of pathological α-synuclein in brain. Brain 136, 1128–1138 (2013).
Osterberg, V. R. et al. Progressive aggregation of α-synuclein and selective degeneration of Lewy inclusion-bearing neurons in a mouse model of parkinsonism. Cell Rep. 10, 1252–1260 (2015).
Peelaerts, W. et al. α-Synuclein strains cause distinct synucleinopathies after local and systemic administration. Nature 522, 340–344 (2015).
Peng, C. et al. Cellular milieu imparts pathological α-synuclein strains in α-synucleinopathies. Nature 557, 558–563 (2018).
Prusiner, S. B. et al. Evidence for α-synuclein prions causing multiple system atrophy in humans with parkinsonism. Proc. Natl Acad. Sci. USA 112, E5308–E5317 (2015).
Tarutani, A., Arai, T., Murayama, S., Hisanaga, S. I. & Hasegawa, M. Potent prion-like behaviors of pathogenic α-synuclein and evaluation of inactivation methods. Acta Neuropathol. Commun. 6, 29 (2018).
Yamasaki, T. R. et al. Parkinson’s disease and multiple system atrophy have distinct α-synuclein seed characteristics. J. Biol. Chem. 294, 1045–1058 (2019).
Lavenir, I. et al. Silver staining (Campbell-Switzer) of neuronal α-synuclein assemblies induced by multiple system atrophy and Parkinson’s disease brain extracts in transgenic mice. Acta Neuropathol. Commun. 7, 148 (2019).
Klingstedt, T. et al. Luminescent conjugated oligothiophenes distinguish between α-synuclein assemblies of Parkinson’s disease and multiple system atrophy. Acta Neuropathol. Commun. 7, 193 (2019).
Strohäker, T. et al. Structural heterogeneity of α-synuclein fibrils amplified from patient brain extracts. Nature Commun. 10, 5535 (2019).
Lau, A. et al. α-Synuclein strains target distinct brain regions and cell types. Nature Neurosci. 23, 21–31 (2020).
Shahnawaz, M. et al. Discriminating α-synuclein strains in Parkinson’s disease and multiple system atrophy. Nature 578, 273–277 (2020).
Ayers, J. I. et al. Different α-synuclein prion strains cause dementia with Lewy bodies and multiple system atrophy. Proc. Natl Acad. Sci. USA 119, e2113489119 (2022).
Spillantini, M. G., Crowther, R. A., Jakes, R., Hasegawa, M. & Goedert, M. α-Synuclein in filamentous inclusions of Lewy bodies from Parkinson’s disease and dementia with Lewy bodies. Proc. Natl Acad. Sci. USA 95, 6469–6473 (1998).
Crowther, R. A., Daniel, S. E. & Goedert, M. Characterisation of isolated α-synuclein filaments from substantia nigra of Parkinson’s disease brain. Neurosci. Lett. 292, 128–130 (2000).
Holec, S. A. M., Liu, S. L. & Woerman, A. L. Consequences of variability in α-synuclein fibril structure on strain biology. Acta Neuropathol. 143, 311–330 (2022).
Zhao, K. et al. Parkinson’s disease-related phosphorylation at Tyr39 rearranges α-synuclein amyloid fibril structure revealed by cryo-EM. Proc. Natl Acad. Sci. USA 117, 20305–20315 (2020).
McGlinchey, R. P., Ni, X., Shadish, J. A., Jiang, J. & Lee, J. C. The N-terminus of α-synuclein dictates fibril formation. Proc. Natl Acad. Sci. USA 118, e2023487118 (2021).
Guerrero-Ferreira, R. et al. Two new polymorphic structures of human full-length alpha-synuclein fibrils solved by cryo-electron microscopy. eLife 8, e48907 (2019).
Shi, Y. et al. Structure-based classification of tauopathies. Nature 598, 359–363 (2021).
Tsuboi, Y. & Dickson, D. W. Dementia with Lewy bodies and Parkinson’s disease with dementia: are they different? Parkinsonism Relat. Disord. 11, S47–S51 (2005).
Compta, Y. et al. Lewy- and Alzheimer-type pathologies in Parkinson’s disease dementia: which is more important? Brain 134, 1493–1505 (2011).
Braak, H. & Del Tredici, K. Neuroanatomy and pathology of sporadic Parkinson’s disease. Adv. Anat. Embryol. Cell Biol. 201, 1–119 (2009).
Poggiolini, I. et al. Diagnostic value of cerebrospinal fluid alpha-synuclein seed quantification in synucleinopathies. Brain 145, 584–595 (2022).
Candelise, N. et al. Seeding variability of different alpha-synuclein strains in synucleinopathies. Ann. Neurol. 85, 691–703 (2019).
Van der Perren, A. et al. The structural differences between patient-derived α-synuclein strains dictate characteristics of Parkinson’s disease, multiple system atrophy and dementia with Lewy bodies. Acta Neuropathol. 139, 977–1000 (2020).
Lövestam, S. et al. Seeded assembly in vitro does not replicate the structures of α-synuclein filaments from multiple system atrophy. FEBS Open Bio 11, 999–1013 (2021).
Burger, D., Fenyi, A., Bousset, L., Stahlberg, H. & Melki, R. Cryo-EM structure of alpha-synuclein fibrils amplified by PMCA from PD and MSA patient brains. Preprint at bioRxiv https://doi.org/10.1101/2021.07.08.451588 (2021).
Frieg, B. et al. α-Synuclein polymorphism determines oligodendroglial dysfunction. Preprint at bioRxiv https://doi.org/10.1101/2021.07.09.451731 (2021).
Sokratian, A. et al. Structural and functional landscape of α-synuclein fibril conformations amplified from cerebrospinal fluid. Preprint at bioRxiv https://doi.org/10.1101/2022.07.13.499896 (2022).
Spillantini, M. G. et al. Filamentous α-synuclein inclusions link multiple system atrophy with Parkinson’s disease and dementia with Lewy bodies. Neurosci. Lett. 251, 205–208 (1998).
Woerman, A. L. et al. Familial Parkinson’s point mutation abolishes multiple system atrophy prion replication. Proc. Natl Acad. Sci. USA 115, 409–414 (2018).
Lövestam, S. et al. Assembly of recombinant tau into filaments identical to those of Alzheimer’s disease and chronic traumatic encephalopathy. eLife 11, e76494 (2022).
Schweighauser, M. et al. Age-dependent formation of TMEM106B amyloid filaments in human brains. Nature 605, 310–314 (2022).
Goedert, M., Spillantini, M. G., Cairns, N. J. & Crowther, R. A. Tau proteins of Alzheimer paired helical filaments: abnormal phosphorylation of all six brain isoforms. Neuron 8, 159–168 (1992).
Giasson, B. I. et al. A panel of epitope-specific antibodies detects protein domains distributes throughout human α-synuclein in Lewy bodies of Parkinson’s disease. J. Neurosci. Res. 59, 528–533 (2000).
Perrin, R. J. et al. Epitope mapping and specificity of the anti-alpha synuclein monoclonal antibody Syn-1 in mouse brain and cultured cell lines. Neurosci. Lett. 349, 133–135 (2003).
Spina, S. et al. The tauopathy associated with mutation +3 in intron 10 of Tau: characterization of the MSTD family. Brain 131, 72–89 (2008).
Zivanov, J., Nakane, T. & Scheres, S. H. W. A Bayesian approach to beam-induced motion correction in cryo-EM single particle analysis. IUCrJ 6, 5–17 (2019).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J.Struct. Biol. 192, 216–221 (2015).
He, S. & Scheres, S. H. W. Helical reconstruction in RELION. J. Struct. Biol. 198, 163–176 (2017).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELON-3. eLife 7, e42166 (2018).
Scheres, S. H. W. Amyloid structure determination in RELION-3.1. Acta Crystallogr. D 76, 94–101 (2020).
Zivanov, J., Nakane, T. & Scheres, S. H. W. Estimation of higher-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ 7, 253–267 (2020).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Casañal, A. et al. Current developments in Coot for macromolecular model building of electron cryo-microscopy and crystallographic data. Protein Sci. 29, 1069–1078 (2020).
Yamashita, K., Palmer, C. M., Burnley, T. & Murshudov, G. N. Cryo-EM single-particle structure refinement and map calculation using Servalcat. Acta Crystallogr. D 77, 1282–1291 (2021).
Acknowledgements
We thank the patients’ families for donating brain tissues, T. Darling and J. Grimmett for help with high-performance computing and the EM facility of the Medical Research Council (MRC) Laboratory of Molecular Biology for help with cryo-EM data acquisition. We thank H. Braak, R.A. Crowther, K. Del Tredici, S. Lövestam, W. Poewe, M.G. Spillantini and E. Tolosa for helpful discussions. We acknowledge Diamond Light Source for access and support of the cryo-EM facilities at the UK’s Electron Bio-imaging Centre (under proposal bi23268), funded by the Wellcome Trust, the MRC and the Biotechnology and Biological Sciences Research Council (BBSRC). This work was supported by the MRC (MC_UP_A025_1013 to S.H.W.S. and MC_U105184291 to M.G.). T.L. holds an Alzheimer’s Research UK Senior Fellowship. T.R. is supported by the National Institute for Health Research Queen Square Biomedical Research Unit in Dementia. The Queen Square Brain Bank is supported by the Rita Lila Weston Institute for Neurological Studies. This work was also supported by the Japan Agency for Science and Technology (CREST) (JPMJCR18H3 to M.H.), the Japan Agency for Medical Research and Development (AMED) (JP20dm0207072 to M.H.), the US National Institutes of Health (P30-AG010133, U01-NS110437 and RF1-AG071177, to R.V. and B.G., and R01NS037167, to T.F.) and the Department of Pathology and Laboratory Medicine, Indiana University School of Medicine (to R.V. and B.G.).
Author information
Authors and Affiliations
Contributions
P.W.C., Y. Saito, T.F., T.T.W., K.H., S.M., T.R., B.G., M.H. and T.L. identified patients and performed neuropathology. Y.Y., H.J.G., R.V. and M.H. performed analysis of brain samples. Y.Y., Y. Shi, M.S., X.Z. and A.K. collected cryo-EM data. Y.Y., Y. Shi, M.S., A.G.M. and S.H.W.S. analysed cryo-EM data. Y.Y. performed immunoblot analysis. B.G., M.H. and T.L. performed immunohistochemistry. S.H.W.S. and M.G. supervised the project. All authors contributed to the writing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
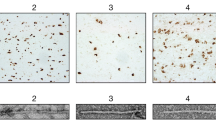
Extended Data Fig. 1 Immunostaining of α-synuclein inclusions.
Sections from brain regions contralateral to those used for cryo-EM structure determination were stained with monoclonal antibody Syn1 (1:1,000). (a), Cingulate cortex from PD; (b), Cingulate cortex from PDD1; (c), Cingulate cortex from PDD2; (d), Frontal cortex from DLB1; (e), Frontal cortex from DLB2; (f), Cingulate cortex from DLB3. Scale bars: a-c, f, 100 μm; d,e, 50 μm.
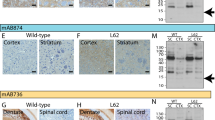
Extended Data Fig. 2 Negative-stain immunoelectron microscopy and immunoblotting of sarkosyl-insoluble material.
PER4 was used at 1:50 in (a–c). (a), PD (Cingulate cortex); (b), PDD1 (Cingulate cortex); (c), DLB3 (Cingulate cortex); Syn303, Syn1 and PER4 were used at 1:4,000 in (d–f). The brain regions used for cryo-EM were also used for immunoblotting. The arrow points to the position of monomeric α-synuclein.
Extended Data Fig. 3 Cryo-EM maps, cryo-EM images and resolution estimates.
(a), α-Synuclein filaments (blue arrows) from PDD1. Scale bars, 50 nm. (b), Projection features of Lewy filament. Scale bars, 5 nm. (c), Zoomed-in view of the main chain showing density of the oxygen atoms. (d), Fourier shell correlation (FSC) curves for cryo-EM maps are shown in black; for the final refined atomic model against the final cryo-EM map in red; for the atomic model refined in the first half map against that half map in blue; for the refined atomic model in the first half map against the other half map in yellow. (e), Side view of α-synuclein filaments with the Lewy fold.
Extended Data Fig. 4 Twisted and untwisted filaments in 2D class averages and projections.
(a,c,e,g), 2D class averages without twist for case 1 of PDD; (b,d,f,h) projections of untwisted models with the Lewy fold, rotated by 0, 220, 110 and 310 degrees, respectively, along the first Euler angle (rot). Box size, 640 Å. (i–p) Projections of untwisted models of the type 3 filaments from seeded assembly with MSA brain (EMD-12269, PDB 7NCK), rotated by 0, 22.5, 45, 67.5, 90, 112.5, 135 and 157.5 degrees, respectively. (q–x) Projections of untwisted models with the MSA type IIA-B folds (EMD-10651, PDB 6XYP), rotated by 0, 22.5, 45, 67.5, 90, 112.5, 135 and 157.5 degrees, respectively.
Extended Data Fig. 5 Twisted and untwisted segments coexist within filaments.
(a,b,c), Micrographs of filaments with untwisted and twisted segments. Blue indicates segments that contributed to twisted 2D class averages; red indicates segments that contributed to untwisted 2D class averages.
Extended Data Fig. 6 Comparison of the Lewy fold with structures of α-synuclein filaments from human brains or assembled from recombinant proteins.
(a), Ribbon plot of the Lewy fold; the protein chain is coloured as in Fig. 2. Highlighted by red, orange, yellow, green, blue and purple areas are substructures that are individually shared with other filament structures. These local similarities are indicated with the same coloured areas and the overlays of the corresponding substructures are shown in sticks on the following panels (b-f). (b), Common core structure of MSA Type I and Type II filaments (made of PFIA/IIA 14–47 and PFIB/IIB 41–99), with a shared substructure highlighted in yellow. (c), pY39 α-synuclein protofilament (PDB:6L1T) with two different substructures highlighted in orange and green. (d), N-terminally truncated α-synuclein (40–140) dimeric filament (PDB:7LC9), with two different substructures in its protofilaments, highlighted in blue and yellow. Residue numbers with apostrophes indicate those from the other protofilament. (e), Polymorph 2a filament (PDB:6SSX), with two substructures highlighted in purple and orange. (f), Polymorph 1a filament (PDB:6H6B) contains yellow-coloured substructures in its protofilaments and a red-coloured substructure in their dimeric interface.
Extended Data Fig. 7 Zoomed-in cryo-EM densities around Y39 of α-synuclein in various folds.
(a), model and cryo-EM densities of the Lewy fold. Models are shown as sticks in green; densities as grey mesh. (b), model and cryo-EM densities of the MSA type I fold (6XYO, EMD-10650). Models are shown as sticks in yellow. (c), model and cryo-EM densities of α-synuclein filaments with phosphorylated Y39 (6L1T, EMD-0801). Models are shown as sticks in blue.
Supplementary information
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, Y., Shi, Y., Schweighauser, M. et al. Structures of α-synuclein filaments from human brains with Lewy pathology. Nature 610, 791–795 (2022). https://doi.org/10.1038/s41586-022-05319-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-05319-3
This article is cited by
-
Misfolded protein oligomers: mechanisms of formation, cytotoxic effects, and pharmacological approaches against protein misfolding diseases
Molecular Neurodegeneration (2024)
-
Disease-specific tau filaments assemble via polymorphic intermediates
Nature (2024)
-
The molecular basis for cellular function of intrinsically disordered protein regions
Nature Reviews Molecular Cell Biology (2024)
-
Phosphorylation and O-GlcNAcylation at the same α-synuclein site generate distinct fibril structures
Nature Communications (2024)
-
RNA modulates hnRNPA1A amyloid formation mediated by biomolecular condensates
Nature Chemistry (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.