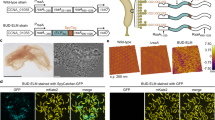
Abstract
Advancing the spontaneous bottom-up construction of artificial cells with high organizational complexity and diverse functionality remains an unresolved issue at the interface between living and non-living matter1,2,3,4. Here, to address this challenge, we developed a living material assembly process based on the capture and on-site processing of spatially segregated bacterial colonies within individual coacervate microdroplets for the endogenous construction of membrane-bounded, molecularly crowded, and compositionally, structurally and morphologically complex synthetic cells. The bacteriogenic protocells inherit diverse biological components, exhibit multifunctional cytomimetic properties and can be endogenously remodelled to include a spatially partitioned DNA–histone nucleus-like condensate, membranized water vacuoles and a three-dimensional network of F-actin proto-cytoskeletal filaments. The ensemble is biochemically energized by ATP production derived from implanted live Escherichia coli cells to produce a cellular bionic system with amoeba-like external morphology and integrated life-like properties. Our results demonstrate a bacteriogenic strategy for the bottom-up construction of functional protoliving microdevices and provide opportunities for the fabrication of new synthetic cell modules and augmented living/synthetic cell constructs with potential applications in engineered synthetic biology and biotechnology.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data supporting the results and conclusions are available within this paper and the Supplementary Information. Source data are provided with this paper.
References
van Stevendaal, M. H. M. E., van Hest, J. C. M. & Mason, A. F. Functional interactions between bottom-up synthetic cells and living matter for biomedical applications. ChemSystemsChem 3, e2100009 (2021).
Jeong, S., Nguyen, H. T., Kim, C. H., Ly, M. N. & Shin, K. Toward artificial cells: novel advances in energy conversion and cellular motility. Adv. Funct. Mater. 30, 1907182 (2020).
Toparlak, O. D. & Mansy, S. S. Progress in synthesizing protocells. Exp. Biol. Med. 244, 304–313 (2018).
Yewdall, N. A., Mason, A. F. & van Hest, J. C. M. The hallmarks of living systems: towards creating artificial cells. Interface Focus 8, 20180023 (2018).
Adamala, K. P., Martin-Alarcon, D. A., Guthrie-Honea, K. R. & Boyden, E. S. Engineering genetic circuit interactions within and between synthetic minimal cells. Nat. Chem. 9, 431–439 (2017).
Deng, N.-N., Yelleswarapu, M., Zheng, L. & Huck, W. T. S. Microfluidic assembly of monodisperse vesosomes as artificial cell models. JACS 139, 587–590 (2017).
Weiss, M. et al. Sequential bottom-up assembly of mechanically stabilized synthetic cells by microfluidics. Nat. Mater. 17, 89–96 (2018).
Huang, X. et al. Interfacial assembly of protein–polymer nano-conjugates into stimulus-responsive biomimetic protocells. Nat. Commun. 4, 2239 (2013).
Li, M., Harbron, R. L., Weaver, J. V. M., Binks, B. P. & Mann, S. Electrostatically gated membrane permeability in inorganic protocells. Nat. Chem. 5, 529–536 (2013).
Marguet, M., Bonduelle, C. & Lecommandoux, S. Multicompartmentalized polymeric systems: towards biomimetic cellular structure and function. Chem. Soc. Rev. 42, 512–529 (2013).
Niederholtmeyer, H., Chaggan, C. & Devaraj, N. K. Communication and quorum sensing in non-living mimics of eukaryotic cells. Nat. Commun. 9, 5027 (2018).
Kumar, B. V. V. S. P., Patil, A. J. & Mann, S. Enzyme-powered motility in buoyant organoclay/DNA protocells. Nat. Chem. 10, 1154–1163 (2018).
Mukwaya, V. et al. Lectin-glycan-mediated nanoparticle docking as a step toward programmable membrane catalysis and adhesion in synthetic protocells. ACS Nano 14, 7899–7910 (2020).
Dou, H. et al. Higher-order assembly of crystalline cylindrical micelles into membrane-extendable colloidosomes. Nat. Commun. 8, 426 (2017).
Martin, N. Dynamic synthetic cells based on liquid–liquid phase separation. ChemBioChem 20, 2553–2568 (2019).
Zhang, Y. et al. Giant coacervate vesicles as an integrated approach to cytomimetic modeling. JACS 143, 2866–2874 (2021).
Tang, T.-Y. D. et al. Fatty acid membrane assembly on coacervate microdroplets as a step towards a hybrid protocell model. Nat. Chem. 6, 527–533 (2014).
Koga, S., Williams, D. S., Perriman, A. W. & Mann, S. Peptide–nucleotide microdroplets as a step towards a membrane-free protocell model. Nat. Chem. 3, 720–724 (2011).
Tang, T.-Y. D., van Swaay, D., deMello, A., Ross Anderson, J. L. & Mann, S. In vitro gene expression within membrane-free coacervate protocells. Chem. Commun. 51, 11429–11432 (2015).
Li, M., Green, D. C., Anderson, J. L. R., Binks, B. P. & Mann, S. In vitro gene expression and enzyme catalysis in bio-inorganic protocells. Chem. Sci. 2, 1739–1745 (2011).
Küchler, A., Yoshimoto, M., Luginbühl, S., Mavelli, F. & Walde, P. Enzymatic reactions in confined environments. Nat. Nanotechnol. 11, 409–420 (2016).
Strulson, C. A., Molden, R. C., Keating, C. D. & Bevilacqua, P. C. RNA catalysis through compartmentalization. Nat. Chem. 4, 941–946 (2012).
Drobot, B. et al. Compartmentalised RNA catalysis in membrane-free coacervate protocells. Nat. Commun. 9, 3643 (2018).
Faust, J. E., Yang, P.-Y. & Huang, H. W. Action of antimicrobial peptides on bacterial and lipid membranes: a direct comparison. Biophys. J. 112, 1663–1672 (2017).
Gray, L. R., Tompkins, S. C. & Taylor, E. B. Regulation of pyruvate metabolism and human disease. Cell. Mol. Life Sci. 71, 2577–2604 (2014).
Silverman, A. D., Karim, A. S. & Jewett, M. C. Cell-free gene expression: an expanded repertoire of applications. Nat. Rev. Genet. 21, 151–170 (2020).
Lee, K. Y. et al. Photosynthetic artificial organelles sustain and control ATP-dependent reactions in a protocellular system. Nat. Biotechnol. 36, 530–535 (2018).
Fung, B. M. & Eyob, E. The effect of ATP concentration on the rate of actin polymerization. Arch. Biochem. Biophys. 220, 370–378 (1983).
Nakashima, K. K., Baaij, J. F. & Spruijt, E. Reversible generation of coacervate droplets in an enzymatic network. Soft Matter 14, 361–367 (2018).
Mempin, R. et al. Release of extracellular ATP by bacteria during growth. BMC Microbiol. 13, 301 (2013).
Alvarez, C. L. et al. Dynamic regulation of extracellular ATP in Escherichia coli. Biochem. J. 474, 1395–1416 (2017).
Donau, C. et al. Active coacervate droplets as a model for membraneless organelles and protocells. Nat. Commun. 11, 5167 (2020).
Moreau, N. G., Martin, N., Gobbo, P., Tang, T. Y. D. & Mann, S. Spontaneous membrane-less multi-compartmentalization via aqueous two-phase separation in complex coacervate micro-droplets. Chem. Commun. 56, 12717–12720 (2020).
Skruzny, M. et al. Molecular basis for coupling the plasma membrane to the actin cytoskeleton during clathrin-mediated endocytosis. Proc. Natl Acad. Sci. USA 109, E2533–E2542 (2012).
Bosnea, L. A., Moschakis, T. & Biliaderis, C. G. Complex coacervation as a novel microencapsulation technique to improve viability of probiotics under different stresses. Food Bioprocess Technol. 7, 2767–2781 (2014).
Florea, M. et al. Engineering control of bacterial cellulose production using a genetic toolkit and a new cellulose-producing strain. Proc. Natl Acad. Sci. USA 113, E3431–E3440 (2016).
Linger, J. G., Adney, W. S. & Darzins, A. Heterologous expression and extracellular secretion of cellulolytic enzymes by Zymomonas mobilis. Appl. Environ. Microbiol. 76, 6360–6369 (2010).
Thakur, S., Weir, B. S. & Guttman, D. S. Phytopathogen genome announcement: draft genome sequences of 62 Pseudomonas syringae type and pathotype strains. Mol. Plant Microbe Interact. 29, 243–246 (2016).
Costales, M. G. et al. A designed small molecule inhibitor of a non-coding RNA sensitizes HER2 negative cancers to herceptin. J. Am. Chem. Soc. 141, 2960–2974 (2019).
Wiechelman, K. J., Braun, R. D. & Fitzpatrick, J. D. Investigation of the bicinchoninic acid protein assay: identification of the groups responsible for color formation. Anal. Biochem. 175, 231–237 (1988).
Drescher, H. K. et al. l-Selectin/CD62L is a key driver of non-alcoholic steatohepatitis in mice and men. Cells 9, 1106 (2020).
Jung, Y. et al. Metabolic signature genes associated with susceptibility to pyruvate kinase, muscle type 2 gene ablation in cancer cells. Mol. Cells 35, 335–341 (2013).
Chiu, J., March, P. E., Lee, R. & Tillett, D. Site-directed, ligase-independent mutagenesis (SLIM): a single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Res. 32, e174 (2004).
Berkson, J. Application of the logistic function to bio-assay. J. Am. Stat. Assoc. 39, 357–365 (1944).
Wittrup, K. D. & Bailey, J. E. A single-cell assay of β-galactosidase activity in Saccharomyces cerevisiae. Cytometry 9, 394–404 (1988).
Spoelstra, W. K., van der Sluis, E. O., Dogterom, M. & Reese, L. Nonspherical coacervate shapes in an enzyme-driven active system. Langmuir 36, 1956–1964 (2020).
Acknowledgements
We thank Y. Takebayashi and J. Spencer for help with bacterial cultures; H. Sun, C. Berger-Schaffitzel and E. Bragginton for help with gel electrophoresis and western blot analysis; and A. Coutable and J. L. R. Anderson for providing the plasmid pEXP5-NT/deGFP. C.X. was funded by a Marie Curie Skłodowska Postdoctoral Fellowship (8082 H2020 PROTOBAC ERC 670 837197) and the BBSRC (BB/P017320/1). N.M. and S.M. were funded by the ERC Advanced Grant Scheme (EC-2016-674 ADG 740235).
Author information
Authors and Affiliations
Contributions
C.X., M.L. and S.M. conceived the experiments. C.X. and N.M. performed the experiments. C.X. and M.L. undertook the data analysis. C.X., M.L. and S.M. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Distribution and viability of co-captured bacterial colonies in coacervate microdroplets.
a–h, Confocal microscopy images (a,c,e,g) and corresponding size distributions (b,d,f,h) of as-prepared PDDA/ATP coacervate droplets (Sul-RhB stained, red fluorescence) (a,b), coacervate droplets (Sul-RhB stained, red fluorescence) with captured E. coli cells (SYTO-9 stained, green fluorescence) (c,d), captured PAO1 cells (SYTO-9 stained, green fluorescence) (e,f), or co-captured E. coli (eGFP-expressed, green fluorescence) and PAO1 (mcherry-expressed, red fluorescence) cells (g,h). i-l, 2D and 3D confocal fluorescence microscopy images of single PDDA/ATP coacervate droplets after incubation with E. coli eGFP (i,j) or P. aeruginosa mCherry (k,l). Expression of eGFP (green fluorescence) or mCherry (red fluorescence) is used to locate the bacteria as dense clusters within the droplet interior (E. coli, i) or as a thin outer shell on the surface of the droplet (P. aeruginosa, k). Live and dead bacterial cells are stained with DNA dyes SYTO-9 (green, membrane permeable) and PI (red, membrane impermeable), respectively. Note the prevalence of living cells associated with both the interior and surface of the coacervate droplets. m–n, FACS 2D gated data (m) and time-dependent plot profiles (n) of live/dead E. coli BL21(DE3) cell population/numbers in PDDA/ATP coacervate droplets after incubation with LB broth. Green dots correspond to live bacteria (stained by SYTO-9); red dots correspond to dead bacteria (stained by PI). o, Plot showing time-dependent decrease in percentage of live E. coli cells within PDDA/ATP coacervate after incubation with LB broth. Number of experiments in n, o, n = 3; error bars, standard deviations. p, Graphic showing transfer of PDDA/ATP coacervate droplets containing sequestered live E. coli BL21(DE3) cells into a petri dish. q, Confocal fluorescence and brightfield microscopy images showing live (green fluorescence)/dead (red fluorescence) staining for E. coli BL21 (DE3) cells in a single PDDA/ATP coacervate after being transferred to a petri dish and cultured for 3 h. The bacteria remain viable within the coacervate droplets under these conditions. r,s Confocal fluorescence and brightfield microscopy images of E. coli Rosetta(DE3) cells with a IPTG-sensitive eGFP plasmid and sequestered within single PDDA/ATP coacervate in the absence (r) and presence (s) of IPTG. Gene expression leading to protein synthesis and folding is only observed in (s). All scale bars, 5 μm.
Extended Data Fig. 2 Construction of bacteriogenic protocells by stepwise in situ lysis of co-captured bacteria populations in coacervate microdroplets.
a, Schematic representation showing disassembly and release of E. coli and/or P. aeruginosa cell membrane and cytoplasmic components via a stepwise lysis procedure using lysozyme and melittin. E. coli and P. aeruginosa are gram-negative bacteria. The procedure is also applicable to gram-positive bacteria such as S. aureus. b–d, Confocal fluorescence microscopy images of PDDA/ATP coacervate droplets containing binary populations of E. coli eGFP and P. aeruginosa mCherry cells before lysis (b), after lysis of only P. aeruginosa mCherry (c), and after lysis of both bacteria (d). Before lysis, populations of E. coli and P. aeruginosa cells are spatially separated between the interior and surface regions of the coacervate droplets, respectively (b; left image, eGFP(green)/mCherry (red) overlay; centre, eGFP; right, mCherry). Lysis of the surface-adsorbed P. aeruginosa population results in coacervate droplets comprising a smooth and continuous shell of phospholipid bilayer membrane fragments derived from the dead P. aeruginosa cells (red fluorescence ring, DilC18 cytoplasmic membrane staining) along with an interior containing high numbers of live E. coli cells (c). Lysis of both bacterial populations disrupts all the cells (DilC18 staining throughout) and releases eGFP into the coacervate matrix to produce bacteriogenic protocells (d). All scale bars, 5 μm. e, Confocal fluorescence microscopy images of a single PDDA/ATP coacervate droplet recorded after capture of E. coli cells and treatment with lysozyme and melittin. Lysis of the E. coli cells releases membrane components (left, DilC18 lipid staining, red fluorescent aggregates), expressed proteins (upper right, eGFP, green fluorescence) and DNA (lower right, SYTO-9/PI staining, red fluorescence) into the coacervate interior. All scale bars, 5 μm. f, As for (e) but for captured and lysed PAO1 cells. Membrane fragments are redistributed around the droplet surface to produce a continuous outer shell (left, DilC18 lipid staining, red fluorescence), and release of low levels of expressed proteins (upper right, m-Cherry, red fluorescence) and DNA (lower right, SYTO-9/PI staining, red fluorescence) into the coacervate matrix. All scale bars, 5 μm. g, Concentrations of eGFP (green column), mCherry (red column) and DNA (blue column) in a population of coacervate droplets after complete lysis of captured E. coli-eGFP or PAO1-mCherry cells. Numbers of samples for tests: n = 3. h, i. Low (left) and high (right) magnification scanning electron micrographs of freeze dried single PDDA/ATP coacervate droplets before (h) and after (i) lysis of captured E. coli and PAO1 cells; the bacteriogenic protocell produced after disassembly of the surface attached PAO1 cells is enclosed by a shell of disordered cytomembrane fragments.
Extended Data Fig. 3 Enzyme activity (β-gal) and glycolysis in bacteriogenic protocells.
a,b, 3D (right) and 2D (left) confocal fluorescence microscopy images of a single bacteriogenic protocell (a) and single coacervate droplet with co-captured non-lysed bacteria (b) 10 min after addition of RBG. β-gal activity is observed only after lysis and construction of the protocells. Typically, the final red fluorescence intensity of the bacteriogenic protocells was about 10 times higher than that of the non-lysed bacteria-loaded droplets. β-gal activity arises from the lysis of E. coli Rosetta (strain DE3) cells, which along with PAO1 cells were used for protocell construction in place of the non-β-gal-active E. coli strain BL21. c, Time-dependent fluorescence microscopy images of a single coacervate droplet with co-captured E. coli Rosetta (strain DE3) and PAO1 cells after addition of RBG showing no apparent β-galactosidase-mediated production and sequestration of resorufin (red fluorescence). The absence of enzyme activity arises from the cell membrane impermeability of RBG. d, Time-dependent fluorescence profiles of RBG reactions with E. coli Rosetta cells (black line) or lysed E. coli Rosetta cells (red line). Ex: 560 nm, Em: 594 nm. The absence of enzyme activity arises from the cell membrane impermeability of RBG45. e, Photographs of the sample well holder containing various samples after 30 min exposure to LB broth and assayed for glycolysis via conversion of pyruvate to l-lactate and formation of the reduced purple dye formazan; native coacervate droplets (i), bacteriogenic protocells (ii), bacterial lysate (iii), E. coli cells and PAO1 (2: 1; OD = 1, iv). Lysate and protocells were generated from the same bacterial concentrations as used in the cell sample. All scale bars, 5 μm.
Extended Data Fig. 4 In situ condensation and spatial localization of bacterial DNA within bacteriogenic protocells.
a, Time-dependent series of confocal microscopy images showing liquid-liquid phase separation and formation of a single DNA-enriched droplet in a PDDA/ATP/histone/DNA coacervate micro-droplet after addition of FITC-CM-dextran (70 kDa). DNA and CM-dextran are labelled with DAPI (blue fluorescence) and FITC (green fluorescence), respectively. CM-dextran is excluded from the DNA-enriched sub-compartment. b, 3D confocal fluorescence microscopy images of a single PDDA/ATP/histone/DNA (wt ratio; 3.2: 5: 2: 1) coacervate droplet after addition of FITC-CM-dextran (70 kDa, green fluorescence). Filtered and overlay images are shown. c, Confocal microscopy images of multiphase coacervate droplets showing co-location of DNA (blue fluorescence) and histone (red fluorescence) in the phase-separated sub-compartment. Samples were prepared using mixtures of PDDA/ATP/RITC-histone/DAPI-DNA. Images were recorded 10 min after addition of FITC-CM-dextran (70 kDa). d, Illustration showing cleavage of circular (plasmid) DNA by DNase I to produce linear dsDNA fragments with blunt ends. e, DNA electrophoresis profiles for commercial purified E. coli DNA before (−) and after (+) incubation with DNase I for 30 min in the presence of aqueous MnCl2. Oligonucleotides with molecular weights less than 200bp are produced. f,g, Confocal bright field (BF) and fluorescence microscopy images of PDDA/ATP/histone/E. coli plasmid DNA coacervate droplets after addition of FITC-CM-dextran (70 kDa, green fluorescence) without (f) or with (g) subsequent treatment with DNase1 to produce linear fragments. Phase separation of the circular or linear DNA produces, respectively, sub-micrometre-sized discontinuous aggregates (f) or a single demixed sub-compartment (g) (blue fluorescence, DAPI-stained DNA). One possibility is that differences in histone-DNA binding for the circular and linear variants influences the surface tension of the phase-separated nuclei and their ability to undergo coalescence. h, Time-dependent series of confocal fluorescence microscopy images of a single bacteriogenic protocell before and after addition of CM-dextran showing nucleation of a population of plasmid DNA/histone condensates (red fluorescence) throughout the molecularly crowded interior. The protocells were prepared from histone-containing PDDA/ATP coacervate droplets followed by in situ lysis of co-captured E. coli and PAO1 cells to release the bacterial DNA in situ. DNA is stained with PI. i, Plot showing time-dependent increase in the number of DNA/histone particles nucleated within individual bacteriogenic protocells after addition of CM-dextran at t = 0. Data normalized to protocell cross-sectional area (number/100 μm2) to account for different protocell sizes. Typically, single protocells would contain around 200 particles of the DNA/histone condensates, which remain dispersed in the protocell interior with minimal aggregation. Number of protocells measured, n = 4; errors bars, standard deviations. All scale bars, 5 μm.
Extended Data Fig. 5 Actin assembly in PDDA/ATP or PDDA/UTP coacervate-based bacteriogenic protocells.
a, 3D confocal fluorescence microscopy images showing in situ assembly of short RhB-labelled actin filaments (red fluorescence) in PDDA/ATP coacervate-based bacteriogenic protocells comprising a bacterial DNA-derived phase-separated histone-containing sub-compartment (proto-nucleus, blue fluorescence, (DAPI stained) embedded in a coacervate matrix (proto-cytoplasm, green fluorescence (FITC-CM-dextran)). Low (left side) and high (right side) magnification images of multiple or single protocells are shown. Uptake of RhB-G-actin from the external solution via diffusion through the protocell membrane (not stained) results in ATP-mediated polymerization to produce fibre-like aggregates specifically in the proto-cytoplasm, but not in the proto-nucleus. Fibre growth was inhibited due to the high concentration of ATP (ca. 200 mM) present in the coacervate phase. b–d. Confocal bright field (BF) and fluorescence microscopy images of single PDDA/UTP coacervate droplets after capture of E. coli (b) or P. aeruginosa (c) cells in the droplet interior or at the surface, respectively. In both cases, the prevalence of live bacteria is shown by SYTO-9 staining of DNA (green fluorescence). d, Single PDDA/UTP/histone/DNA coacervate droplet after addition of FITC-CM-dextran (green fluorescence) showing formation of a phase separated DNA/histone-enriched sub-compartment (DAPI stained, blue fluorescence). e, 2D (left) and 3D (right) confocal fluorescence microscopy images showing the distribution of actin (red fluorescence, RhB-labelled) in a single PDDA/UTP coacervate-based bacteriogenic protocell containing a phase-separated DNA/histone sub-compartment (blue fluorescence, (DAPI stained) dispersed in a FITC-CM-dextran-enriched coacervate matrix (green fluorescence). No ATP-generating system is present. As a consequence, actin-rich aggregates, 0.1–1 μm in size (red spots) in place of F- actin filaments are observed. All scale bars, 5 μm.
Extended Data Fig. 6 Viability of implanted E. coli cells in PDDA/UTP coacervate-based bacteriogenic protocells.
a, Confocal fluorescence microscopy images showing live/dead stain for E. coli BL21 (DE3) cells in a single PDDA/UTP coacervate-based bacteriogenic protocell with a single proto-nucleus (blue/red fluorescence, DAPI/PI stained). To inactivate the lysozyme and melittin inside the protocell and in the solution, the protocells were pre-incubated with lysozyme inhibitor NAG3 and melittin inhibitor OBAA for 0.5 h before adding of E. coli. The live E. coli cells were stained by SYTO-9 (green fluorescence), which can penetrate live bacteria, and dead cells were stained by PI (red fluorescence), which can only penetrate dead bacteria and can also stain the DNA in the proto-nucleus. Images showed that almost all of the E. coli cells remained alive in the protocells after 3 h. Scale bar, 5 μm. b,c, FACS 2D gated data (b) and time-dependent plot profiles (c) of live/dead E. coli cell population/numbers in bacteriogenic protocells after incubation with LB broth. Green dots correspond to live bacteria (stained by SYTO-9); red dots correspond to dead bacteria (stained by PI). d, Plot showing time-dependent decrease in percentage of live E. coli cells within bacteriogenic protocells after incubation with LB broth.
Extended Data Fig. 7 Enhanced cytomimetic properties of bacteriogenic protocells energized by implanted live E. coli cells.
a, Time-dependent plots of ATP concentrations produced in bacteriogenic protocells via endogenous bio-generation (entrapped live E. coli cells) over a period of 72 h. The bacterial ATP profiles were dependent on the nutrient levels in the experimental system. Number of samples, n = 3; error bars are standard deviations. b, c, Schematic diagram of the Z’-LYTE biochemical assay for kinase activity detection in protocells. The Z’-LYTE biochemical assay employs a FRET-based, coupled-enzyme format and is based on the differential sensitivity of phosphorylated and non-phosphorylated peptides to proteolytic cleavage. The pentapeptide substrate is labelled with two fluorophores, coumarin (C) and fluorescein (F) —one at each end—that make up a FRET pair. In the primary reaction (b, the kinase reaction), the kinase transfers the γ-phosphate of ATP to a single serine or threonine residue in the synthetic peptide substrate. In the secondary reaction (c, the Development reaction), a site-specific protease (the Development reagent) recognizes and cleaves non-phosphorylated peptides. Phosphorylated peptides exhibit suppressed cleavage by the Development reagent. Cleavage disrupts FRET between the donor (coumarin) and acceptor (fluorescein) fluorophores on the peptide, whereas uncleaved phosphorylated peptides maintain FRET. A ratiometric method, which calculates the ratio (the Emission Ratio = Coumarin Emission (445 nm)/Fluorescein Emission (520 nm) of donor emission to acceptor emission after excitation of the donor fluorophore at 400 nm, quantitates reaction progress. d, The emission ratio (445/520) of protocells with endogenous ATP bio-generation (entrapped live E. coli cells (+), green column, 1.7), with endogenous ATP chemical (enzyme) generation (PK/PEP/ADP pathway, purple column, 2.3) or ATP present in the released bacterial lysate (no live E. coli, grey column, 3.9) after incubation with Z’-LYTE biochemical substrate reagents and developing reagent for 1 h under room temperature. Number of samples, n = 3; error bars are standard deviations. e–j, In vitro translation and transcription in live E. coli-containing bacteriogenic protocells (PDDA/UTP); Confocal fluorescence (e) and bright field (g) microscopy image, 3D fluorescence surface plot (f) and weight percent (%) and of deGFP partitioning (h) in the cytoplasm- and nucleus-like regions in a single hybrid protocell. Scale bars, 5 μm. Gel electrophoresis profiles (i) and western blotting images (j) of extracted proteins obtained after salt-induced disassembly of protocells with (+) or without (−) plasmid pEXP5-NT/deGFP. Arrow in (i, j) shows additional band at 27 kDa that was absent without the plasmid and which stains on exposure to an anti-GFP antibody (chicken) and goat anti-chicken IgY H&L (alkaline phosphatase). k, Glycolysis in ATP-generating E. coli-containing bacteriogenic protocells (PDDA/UTP); time-dependent plot of changes in absorption at 565 nm, final concentration of l-lactate produced (insert, left) and photograph of sample after 30 min (insert, right) associated with endogenous ATP biogeneration. Number of samples, n = 3; error bars are standard deviations. For comparison with protocells containing only released bacterial lysate; see Fig. 3h–j in the main manuscript.
Extended Data Fig. 8 F-actin-mediated structural stability of living/synthetic cell constructs.
a, 3D confocal fluorescence microscopy images of an individual living/synthetic cell construct showing extensive network of F-actin micro-filaments (red fluorescence, RhB-labelled actin) within the cytoplasmic-like space of a bacteriogenic host protocell. The DNA/histone sub-compartment (blue fluorescence, DAPI, DNA stain) and guest E. coli eGFP cells (green fluorescence, expressed GFP) are immobilized within the cytoskeletal-like matrix. Continuous on-site bio-production of ATP from a glucose source and uptake of G-actin give rise to extensive hydrogelation of the protocell interior. Images are recorded 0.5 h after addition of G-actin. b,c, Confocal fluorescence microscopy images recorded at low (b) and high (c) magnification of a single bacteriogenic protocell with entrapped live E. coli cells showing extensive cytoskeletal-like network (red fluorescence, RhB-F-actin filaments) within the cytoplasmic-like space of the bacteriogenic host protocell. The DNA/histone proto-nucleus (blue fluorescence, DAPI DNA stain) and guest E. coli eGFP cells (green fluorescence, expressed GFP) are immobilized within the cytoskeletal-like matrix. Images recorded 2 days after completion of internal hydrogelation. The enlarged image shown in c is taken from the dashed box shown in b. d,e. Confocal fluorescence microscopy of bacteriogenic hybrid protocell with (d) and without (e) F-actin after exposure to 200 mM NaCl (added at t = 6 s (white arrows)). Outer membrane (red fluorescence, Dil stain; PAO1-derived lipids), molecularly crowded proto-cytoplasm (non-fluorescent), proto-cytoskeleton (red fluorescence), membrane-free DNA/histone proto-nucleus (blue fluorescence). The protocells with F-actin (d) showing minimal shrinking under hypertonic conditions, while without F-actin, the protocells are disassembled. All scale bars, 5 μm.
Extended Data Fig. 9 Mechanism of live cell-mediated morphogenesis in bacteriogenic protocells.
a, Confocal fluorescence microscopy image of an individual bacteriogenic protocell with F-actin and endogenous ATP chemical (enzyme) generation (PK/PEP/ADP pathway) after incubation with substrates (PEP/ADP) for 1 day showing no morphological change associated with F-actin or ATP production. b, Confocal fluorescence microscopy image of an individual amoeba-like bacteriogenic protocell with entrapped E. coli eGFP cells after incubation with LB broth for 1 day. Scale bars, 5 μm. c, Percentage of protocells with non-spherical morphologies using a circularity analysis46. Number of samples, n = 3; error bars, standard deviations. d,e Optical images (d) and contact angle plot (e) showing time-dependent increase in wettability and decrease in contact angle for single droplets of a concentrated suspension of living/synthetic constructs mounted on a pegylated glass slide. f, 2D Confocal fluorescence microscopy images (top) and 3D Imaris reconstruction images (bottom) of bacteriogenic protocells with entrapped E. coli eGFP cells showing progressive formation of amoeba-like living/synthetic microscale constructs with retention of internal organization and outer membrane after incubation on a non-modified glass cover slide or plastic cover slide. Data recorded at 1 h and 48 h. Staining: DNA/histone condensate (DAPI, blue); guest live E. coli cells (eGFP, green dots); outer membrane (DilC18, red). Scale bars, 10 μm. g, Confocal microscopy image (left) of an individual bacteriogenic protocell containing entrapped E. coli cells and F-actin after incubation with LB broth for 48 h; the image was recorded in aqueous suspension, DNA staining by DAPI, blue fluorescence, scale bar, 10 μm. h, Confocal fluorescence microscopy image of an individual bacteriogenic protocell with high levels of entrapped E. coli eGFP cells after incubation with LB broth for 1 h. The E. coli-eGFP cells were washed with PBS buffer to remove bacterial secretions before addition to the protocells. Scale bar, 5 μm.
Extended Data Fig. 10 Time-dependent decrease in membrane permeability of living/synthetic constructs.
a-d, Confocal fluorescence microscopy images of single living/synthetic constructs incubated for 48 h followed by addition of various green-fluorescent solutes to the external phase (a), and corresponding uptake ratios (grey-value ratios, FLinterior/FLexterior) (b), showing impermeability of 10 and 40 kDa FITC-dextran. c, as for (a) but after different periods of biological activity followed by addition of 40 kDa dextran. d, Corresponding uptake ratios determined for different incubation times shown in (c); e,f Confocal fluorescence microscopy images (e) and corresponding uptake ratios (grey-value ratios, FLinterior/FLexterior) (f) of single living/synthetic constructs incubated for 1h and 48 h followed by addition of various green-fluorescent solutes (FITC-BSA, Mw = 66 kDa), eGFP (32 kDa) and ssDNA-FAM (8 Da) to the external phase. All scale bars, 5 μm.
Supplementary information
Supplementary Information
Supplementary Figs. 1–11.
Supplementary Video 1
Spontaneous capture of E. coli cells inside PDDA/ATP coacervate microdroplets. Confocal microscopy video (optical and fluorescence overlay) showing a single PDDA/ATP coacervate droplet in the presence of flowing stream of SYTO-9 stained E. coli cells. The bacterial cells readily penetrate the droplet surface and become trapped within the molecularly crowded coacervate matrix. The video runs for 5 min 50 s; real time is shown at the top left.
Supplementary Video 2
Spontaneous capture of PAO1 cells on the surface of PDDA/ATP coacervate microdroplets. Confocal microscopy video (optical and fluorescence overlay) showing a single PDDA/ATP coacervate droplet in the presence of flowing stream of SYTO-9 stained PAO1 cells. The bacterial cells do not penetrate the droplet but remain attached to the droplet surface to produce an outer shell. The video runs for 1 min 26 s; real time is shown at the top left.
Supplementary Video 3
Spontaneous capture of E. coli and PAO1 cells by PDDA/ATP coacervate microdroplets. Confocal microscopy video (optical and fluorescence overlay) showing PDDA/ATP coacervate droplets in the presence of flowing stream of E. coli eGFP (green fluorescence) and PAO1 mCherry (red fluorescence). The E. coli cells are trapped and aggregated inside the droplets while the PAO1 cells attach to the droplet surface to produce a continuous outer shell. The video runs for 1 min 26 s; real time is shown at the top left.
Supplementary Video 4
Enzyme β-gal activity in a single bacteriogenic protocell. Fluorescence microscopy video showing a single PDDA/ATP coacervate droplet loaded with living E. coli Rosetta(DE3) and PAO1 cells before lysis (left) and a single bacteriogenic protocell prepared after bacterial lysis (right); in both cases, resorufin-β-d-galactopyranoside (RBG) was added to the external environment at t = 0. Production of resorufin by β-gal occurs only in the protocell (onset of red fluorescence) due to accessibility of the released enzyme to the added RBG. Real time is shown at the top left of the two panels.
Source data
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, C., Martin, N., Li, M. et al. Living material assembly of bacteriogenic protocells. Nature 609, 1029–1037 (2022). https://doi.org/10.1038/s41586-022-05223-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-05223-w
This article is cited by
-
Present and future of synthetic cell development
Nature Reviews Molecular Cell Biology (2024)
-
Self-assembly of stabilized droplets from liquid–liquid phase separation for higher-order structures and functions
Communications Chemistry (2024)
-
Multicompartmental coacervate-based protocell by spontaneous droplet evaporation
Nature Communications (2024)
-
An inorganic mineral-based protocell with prebiotic radiation fitness
Nature Communications (2023)
-
Multi-compartmental MOF microreactors derived from Pickering double emulsions for chemo-enzymatic cascade catalysis
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.