Abstract
In European and many African, Middle Eastern and southern Asian populations, lactase persistence (LP) is the most strongly selected monogenic trait to have evolved over the past 10,000 years1. Although the selection of LP and the consumption of prehistoric milk must be linked, considerable uncertainty remains concerning their spatiotemporal configuration and specific interactions2,3. Here we provide detailed distributions of milk exploitation across Europe over the past 9,000 years using around 7,000 pottery fat residues from more than 550 archaeological sites. European milk use was widespread from the Neolithic period onwards but varied spatially and temporally in intensity. Notably, LP selection varying with levels of prehistoric milk exploitation is no better at explaining LP allele frequency trajectories than uniform selection since the Neolithic period. In the UK Biobank4,5 cohort of 500,000 contemporary Europeans, LP genotype was only weakly associated with milk consumption and did not show consistent associations with improved fitness or health indicators. This suggests that other reasons for the beneficial effects of LP should be considered for its rapid frequency increase. We propose that lactase non-persistent individuals consumed milk when it became available but, under conditions of famine and/or increased pathogen exposure, this was disadvantageous, driving LP selection in prehistoric Europe. Comparison of model likelihoods indicates that population fluctuations, settlement density and wild animal exploitation—proxies for these drivers—provide better explanations of LP selection than the extent of milk exploitation. These findings offer new perspectives on prehistoric milk exploitation and LP evolution.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Data for running the aDNA analyses are available from https://github.com/ydiekmann/Evershed_Nature_2022. KML files, a summary of archaeological milk residue data, ecological proxy variables and a summary of radiocarbon dates are available from https://github.com/AdrianTimpson/2020-03-03523A. UK Biobank data are available from: https://www.ukbiobank.ac.uk/.
Code availability
R code for running the aDNA analyses is available from https://github.com/ydiekmann/Evershed_Nature_2022. Open-source R Code for running the UK Biobank analyses under MIT license are available from https://github.com/MRCIEU/lp-coevolution. R code for the generation of Figs. 1, 2, 3 and Extended Data Fig. 1 are available from https://github.com/AdrianTimpson/2020-03-03523A.
Change history
28 July 2022
The online PDF was replaced as the last two authors were deleted in error in the original PDF.
30 August 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41586-022-05227-6
References
Sabeti, P. C. et al. Positive natural selection in the human lineage. Science 312, 1614 (2006).
Evershed, R. P. et al. Earliest date for milk use in the Near East and southeastern Europe linked to cattle herding. Nature 455, 528–531 (2008).
Debono Spiteri, C. et al. Regional asynchronicity in dairy production and processing in early farming communities of the northern Mediterranean. Proc. Natl Acad. Sci. USA 113, 13594–13599 (2016).
Collins, R. What makes UK Biobank special? Lancet 379, 1173–1174 (2012).
Allen, N. E., Sudlow, C., Peakman, T. & Collins, R. UK Biobank Data: come and get it. Sci. Transl. Med. 6, 224ed224–224ed224 (2014).
Gerbault, P. et al. Evolution of lactase persistence: an example of human niche construction. Philos. Trans. R. Soc. Lond. B 366, 863–877 (2011).
Food and Agriculture Organization of the United Nations. Crops and Livestock Products (FAOSTAT) http://www.fao.org/faostat/en/#data/QA (accessed 10 November 2021).
Vigne, J.-D. & Helmer, D. Was milk a “secondary product” in the Old World Neolithisation process? Its role in the domestication of cattle, sheep and goats. Anthropozoologica 42, 9–40 (2007).
Roffet-Salque, M., Gillis, R., Evershed, R. P. & Vigne, J.-D. in Hybrid Communities: Biosocial Approaches to Domestication and Other Trans-Species Relationships (eds Stépanoff, C. & Vigne, J.D.) 127–143 (Routledge, 2018).
Gillis, R. et al. Sophisticated cattle dairy husbandry at Borduşani-Popină (Romania, fifth millennium BC): the evidence from complementary analysis of mortality profiles and stable isotopes. World Archaeol. 45, 447–472 (2013).
Ethier, J. et al. Earliest expansion of animal husbandry beyond the Mediterranean zone in the sixth millennium BC. Sci. Rep. 7, 7146 (2017).
Salque, M. et al. Earliest evidence for cheese making in the sixth millennium BC in northern Europe. Nature 493, 522–525 (2013).
Gillis, R. E. et al. The evolution of dual meat and milk cattle husbandry in Linearbandkeramik societies. Proc. R. Soc. B 284, 20170905 (2017).
Balasse, M. & Tresset, A. Early weaning of Neolithic domestic cattle (Bercy, France) revealed by intra-tooth variation in nitrogen isotope ratios. J. Archaeol. Sci. 29, 853–859 (2002).
Whelton, H. L., Roffet-Salque, M., Kotsakis, K., Urem-Kotsou, D. & Evershed, R. P. Strong bias towards carcass product processing at Neolithic settlements in northern Greece revealed through absorbed lipid residues of archaeological pottery. Quat. Int. 496, 127–139 (2018).
Copley, M. S. et al. Direct chemical evidence for widespread dairying in prehistoric Britain. Proc. Natl Acad. Sci. USA 100, 1524–1529 (2003).
Cramp, L. J. E. et al. Immediate replacement of fishing with dairying by the earliest farmers of the northeast Atlantic archipelagos. Proc. R. Soc. B 281, 20132372 (2014).
Smyth, J. & Evershed, R. P. Milking the megafauna: using organic residue analysis to understand early farming practice. Environ. Archaeol. 21, 214–229 (2016).
Charlton, S. et al. New insights into Neolithic milk consumption through proteomic analysis of dental calculus. Archaeol. Anthropol. Sci. 11, 6183–6196 (2019).
Craig, O. E. et al. Ancient lipids reveal continuity in culinary practices across the transition to agriculture in Northern Europe. Proc. Natl Acad. Sci. USA 108, 17910–17915 (2011).
Cramp, L. J. E. et al. Neolithic dairy farming at the extreme of agriculture in Northern Europe. Proc. R. Soc. B 281, 20140819 (2014).
Pääkkönen, M., Holmqvist, E., Bläuer, A., Evershed, R. P. & Asplund, H. Diverse economic patterns in the North Baltic Sea region in the Late Neolithic and Early Metal periods. Eur. J. Archaeol. 23, 4–21 (2019).
Burger, J., Kirchner, M., Bramanti, B., Haak, W. & Thomas, M. G. Absence of the lactase-persistence-associated allele in early Neolithic Europeans. Proc. Natl Acad. Sci. USA 104, 3736–3741 (2007).
Sverrisdóttir, O. Ó. et al. Direct estimates of natural selection in Iberia indicate calcium absorption was not the only driver of lactase persistence in Europe. Mol. Biol. Evol. 31, 975–983 (2014).
Allentoft, M. E. et al. Population genomics of Bronze Age Eurasia. Nature 522, 167–172 (2015).
Haak, W. et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522, 207–211 (2015).
Brace, S. et al. Ancient genomes indicate population replacement in Early Neolithic Britain. Nat. Ecol. Evol. 3, 765–771 (2019).
Burger, J. et al. Low prevalence of lactase persistence in Bronze Age Europe indicates ongoing strong selection over the last 3,000 years. Curr. Biol. 30, 4307–4315 (2020).
Itan, Y., Powell, A., Beaumont, M. A., Burger, J. & Thomas, M. G. The origins of lactase persistence in Europe. PLoS Comput. Biol. 5, e1000491 (2009).
Enattah, N. S. et al. Identification of a variant associated with adult-type hypolactasia. Nat. Genet. 30, 233–237 (2002).
Itan, Y., Jones, B., Ingram, C., Swallow, D. & Thomas, M. A worldwide correlation of lactase persistence phenotype and genotypes. BMC Evol. Biol. 10, 36 (2010).
Flatz, G. & Rotthauwe, H. Lactose nutrition and natural selection. Lancet 302, 76–77 (1973).
Cubas, M. et al. Latitudinal gradient in dairy production with the introduction of farming in Atlantic Europe. Nat. Commun. 11, 2036 (2020).
Klopfstein, S., Currat, M. & Excoffier, L. The fate of mutations surfing on the wave of a range expansion. Mol. Biol. Evol. 23, 482–490 (2006).
Gerbault, P., Moret, C., Currat, M. & Sanchez-Mazas, A. Impact of selection and demography on the diffusion of lactase persistence. PLoS ONE 4, e6369 (2009).
Cook, G. C. & al-Torki, M. T. High intestinal lactase concentrations in adult Arabs in Saudi Arabia. Br. Med. J. 3, 135–136 (1975).
Davey Smith, G. et al. Lactase persistence-related genetic variant: population substructure and health outcomes. Eur. J. Hum. Genet. 17, 357–367 (2009).
Cederlund, A. et al. Lactose in human breast milk an inducer of innate immunity with implications for a role in intestinal homeostasis. PLoS ONE https://doi.org/10.1371/journal.pone.0053876 (2013).
Gibson, P. R. History of the low FODMAP diet. J. Gastroenterol. Hepatol. 32, 5–7 (2017).
Walker, C. & Thomas, M. G. in Lactose: Evolutionary Role, Health Effects, and Applications (eds Paques, M. & Lindner, C.) 1–48 (Elsevier, 2019).
Simoons, F. Primary adult lactose intolerance and the milking habit: a problem in biologic and cultural interrelations. II. A culture historical hypothesis. Dig. Dis. Sci. 15, 695–710 (1970).
McCracken, R. D. Lactase deficiency: an example of dietary evolution. Curr. Anthropol. 12, 479–517 (1971).
Holden, C. & Mace, R. Phylogenetic analysis of the evolution of lactose digestion in adults. Hum. Biol. 69, 605–628 (1997).
Ingram, C. J. E. et al. A novel polymorphism associated with lactose tolerance in Africa: multiple causes for lactase persistence? Hum. Genet. 120, 779–788 (2007).
Tishkoff, S. A. et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat. Genet. 39, 31–40 (2007).
Joslin, S. E. K. et al. Association of the lactase persistence haplotype block with disease risk in populations of European descent. Front. Genet. 11, 1346 (2020).
Dudd, S. N. & Evershed, R. P. Direct demonstration of milk as an element of archaeological economies. Science 282, 1478–1481 (1998).
Dunne, J. et al. First dairying in green Saharan Africa in the fifth millennium BC. Nature 486, 390–394 (2012).
Manning, K. et al. The origins and spread of stock-keeping: the role of cultural and environmental influences on early Neolithic animal exploitation in Europe. Antiquity 87, 1046–1059 (2013).
Shennan, S. et al. Regional population collapse followed initial agriculture booms in mid-Holocene Europe. Nat. Commun. 4, 2486 (2013).
Howe, L. J. et al. Genetic evidence for assortative mating on alcohol consumption in the UK Biobank. Nat. Commun. 10, 5039 (2019).
Rodriguez, S., Gaunt, T. R. & Day, I. N. M. Hardy–Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. Am. J. Epidemiol. 169, 505–514 (2009).
Simmons, J. S., Whayne, T. F., Anderson, G. W. & Horack, H. M. Global Epidemiology: A Geography of Disease and Sanitation Vol. 1 (William Heineman, 1944).
Bai, Z. et al. Global environmental costs of China's thirst for milk. Glob. Change Biol. 24, 2198–2211 (2018).
Simoons, F. Primary adult lactose intolerance and the milking habit: a problem in biological and cultural interrelations. I. Review of the medical research. Dig. Dis. Sci. 14, 819–836 (1969).
Bayless, T. M., Paige, D. M. & Ferry, G. D. Lactose intolerance and milk drinking habits. Gastroenterology 60, 605–608 (1971).
Szilagyi, A., Walker, C. & Thomas, M. G. in Lactose: Evolutionary Role, Health Effects, and Applications (eds Paques, M. & Lindner, C.) 113–153 (Elsevier, 2019).
Mendelian Randomization of Dairy Consumption Working Group. Dairy consumption and body mass index among adults: Mendelian randomization analysis of 184802 individuals from 25 studies. Clin. Chem. 64, 183–191 (2018).
Almon, R., Álvarez-León, E. E. & Serra-Majem, L. Association of the European lactase persistence variant (LCT-13910 C>T polymorphism) with obesity in the Canary Islands. PLoS ONE https://doi.org/10.1371/journal.pone.0043978 (2012).
Hartwig, F. P., Horta, B. L., Davey Smith, G., de Mola, C. L. & Victora, C. G. Association of lactase persistence genotype with milk consumption, obesity and blood pressure: a Mendelian randomization study in the 1982 Pelotas (Brazil) Birth Cohort, with a systematic review and meta-analysis. Int. J. Epidemiol. 45, 1573–1587 (2016).
Qin, L.-Q., He, K. & Xu, J.-Y. Milk consumption and circulating insulin-like growth factor-I level: a systematic literature review. Int. J. Food Sci. Nutr. 60, 330–340 (2009).
Wiley, A. The evolution of lactase persistence: milk consumption, insulin-like growth factor I, and human life-history parameters. Q. Rev. Biol. 93, 319–345 (2018).
Campbell, C. D. et al. Demonstrating stratification in a European American population. Nat. Genet. 37, 868–872 (2005).
Bergholdt, H. K. M., Nordestgaard, B. G., Varbo, A. & Ellervik, C. Milk intake is not associated with ischaemic heart disease in observational or Mendelian randomization analyses in 98,529 Danish adults. Int. J. Epidemiol. 44, 587–603 (2015).
Bocquet-Appel, J.-P. When the world's population took off: the springboard of the Neolithic demographic transition. Science 333, 560–561 (2011).
Loog, L. et al. Estimating mobility using sparse data: application to human genetic variation. Proc. Natl Acad. Sci. USA 114, 12213–12218 (2017).
Vorou, R., Papavassiliou, V. & Tsiodras, S. Emerging zoonoses and vector-borne infections affecting humans in Europe. Epidemiol. Infect. 135, 1231–1247 (2007).
Allen, T. et al. Global hotspots and correlates of emerging zoonotic diseases. Nat. Commun. 8, 1124 (2017).
Rice, A. L., Sacco, L., Hyder, A. & Black, R. E. Malnutrition as an underlying cause of childhood deaths associated with infectious diseases in developing countries. Bull. World Health Org. 78, 1207–1221 (2000).
Colledge, S., Conolly, J., Crema, E. & Shennan, S. Neolithic population crash in northwest Europe associated with agricultural crisis. Quat. Res. 92, 686–707 (2019).
Manning, K. The cultural evolution of Neolithic Europe. EUROEVOL Dataset 2: Zooarchaeological data. J. Open Archaeol. Data 5, e3 (2016).
Chessa, B. et al. Revealing the history of sheep domestication using retrovirus integrations. Science 324, 532 (2009).
Karesh, W. B., Cook, R. A., Bennett, E. L. & Newcomb, J. Wildlife trade and global disease emergence. Emerg. Infect. Dis. 11, 1000 (2005).
Chomel, B. B., Belotto, A. & Meslin, F.-X. Wildlife, exotic pets, and emerging zoonoses. Emerg. Infect. Dis. 13, 6 (2007).
Schibler, J., Jacomet, S., Hüster-Plogmann, H. & Brombacher, C. Economic crash in the 37th and 36th centuries cal. BC in Neolithic lake shore sites in Switzerland. Anthropozoologica 25, 553–570 (1997).
He, Y., Yang, X., Xia, J., Zhao, L. & Yang, Y. Consumption of meat and dairy products in China: a review. Proc. Nutr. Soc. 75, 385–391 (2016).
Mak, V. S. W. Milk Craze: Body, Science, and Hope in China (Univ. of Hawai'i Press, 2021).
Goodrich, J. K. et al. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe 19, 731–743 (2016).
Qin, Y. et al. Combined effects of host genetics and diet on human gut microbiota and incident disease in a single population cohort. Nat. Genet. 54, 134–142 (2022).
Paine, R. R. & Boldsen, J. L. in The Evolution of Human Life History School for Advanced Research (eds Hawkes, K. & Paine, R. R.) 307–330 (School of American Research Press, 2006).
Stackhouse P. W. et al. POWER Release 8.0.1 (with GIS Applications) Methodology (Data Parameters, Sources, & Validation) (NASA, 2018).
Lieberman, M. & Lieberman, D. Lactase deficiency: a genetic mechanism which regulates the time of weaning. Am. Nat. 112, 625–627 (1978).
Ingold, T. Hunters, Pastoralists and Ranchers: Reindeer Economies and Their Transformations (Cambridge Univ. Press, 1980).
Outram, A. K. et al. The earliest horse harnessing and milking. Science 323, 1332–1335 (2009).
Breu, A., Gómez-Bach, A., Heron, C., Rosell-Melé, A. & Molist, M. Variation in pottery use across the Early Neolithic in the Barcelona plain. Archaeol. Anthropol. Sci. 13, 53 (2021).
Brychova, V. et al. Animal exploitation and pottery use during the early LBK phases of the Neolithic site of Bylany (Czech Republic) tracked through lipid residue analysis. Quat. Int. 574, 91–101 (2021).
Carrer, F. et al. Chemical analysis of pottery demonstrates prehistoric origin for high-altitude alpine dairying. PLoS ONE 11, e0151442 (2016).
Casanova, E. et al. Spatial and temporal disparities in human subsistence in the Neolithic Rhineland gateway. J. Archaeol. Sci. 122, 105215 (2020).
Colonese, A. C. et al. The identification of poultry processing in archaeological ceramic vessels using in-situ isotope references for organic residue analysis. J. Archaeol. Sci. 78, 179–192 (2017).
Copley, M. S. et al. Dairying in antiquity. I. Evidence from absorbed lipid residues dating to the British Iron Age. J. Archaeol. Sci. 32, 485–503 (2005).
Copley, M. S., Berstan, R., Straker, V., Payne, S. & Evershed, R. P. Dairying in antiquity. II. Evidence from absorbed lipid residues dating to the British Bronze Age. J. Archaeol. Sci. 32, 505–521 (2005).
Copley, M. S. et al. Dairying in antiquity. III. Evidence from absorbed lipid residues dating to the British Neolithic. J. Archaeol. Sci. 32, 523–546 (2005).
Copley, M. S. & Evershed, R. P. in Building Memories: the Neolithic Cotswold Long Barrow at Ascott-under-Wychwood, Oxfordshire (eds Benson, D. & Whittle, A.) 283–288 (Oxbow Books, 2006).
Courel, B. et al. Organic residue analysis shows sub-regional patterns in the use of pottery by Northern European hunter–gatherers. R. Soc. Open Sci. 7, 192016 (2020).
Craig, O. E. et al. Did the first farmers of central and eastern Europe produce dairy foods? Antiquity 79, 882–894 (2005).
Craig, O. E., Taylor, G., Mulville, J., Collins, M. J. & Parker Pearson, M. The identification of prehistoric dairying activities in the Western Isles of Scotland: an integrated biomolecular approach. J. Archaeol. Sci. 32, 91–103 (2005).
Craig, O. E. et al. Molecular and isotopic demonstration of the processing of aquatic products in Northern European Prehistoric pottery. Archaeometry 49, 135–152 (2007).
Craig, O. in Archaeology Meets science: Biomolecular Investigations in Bronze Age Greece (eds Tzedakis, Y. et. al.) 121–124 (Oxbow Books, 2008).
Craig, O. E. et al. Feeding Stonehenge: cuisine and consumption at the Late Neolithic site of Durrington Walls. Antiquity 89, 1096–1109 (2015).
Craig-Atkins, E. et al. The dietary impact of the Norman Conquest: a multiproxy archaeological investigation of Oxford, UK. PLoS ONE 15, e0235005 (2020).
Cramp, L. J. E., Evershed, R. P. & Eckardt, H. What was a mortarium used for? Organic residues and cultural change in Iron Age and Roman Britain. Antiquity 85, 1339–1352 (2011).
Cramp, L. J. E. et al. Regional diversity in subsistence among early farmers in Southeast Europe revealed by archaeological organic residues. Proc. R. Soc. B 286, 20182347 (2019).
Cramp, L. J. E., Król, D., Rutter, M., Heyd, V. M. & Pospieszny, L. Analiza pozostałości organicznych z ceramiki kultury rzucewskiej z Rzucewa. Pomorania Antiqua XXVIII, 245–259 (2019).
Demirci, Ö., Lucquin, A., Craig, O. E. & Raemaekers, D. C. M. First lipid residue analysis of Early Neolithic pottery from Swifterbant (the Netherlands, ca. 4300–4000 BC). Archaeol. Anthropol. Sci. 12, 105 (2020).
Demirci, Ö., Lucquin, A., Çakırlar, C., Craig, O. E. & Raemaekers, D. C. M. Lipid residue analysis on Swifterbant pottery (c. 5000–3800 cal bc) in the Lower Rhine–Meuse area (the Netherlands) and its implications for human–animal interactions in relation to the Neolithisation process. J. Archaeol. Sci. Rep. 36, 102812 (2021).
Dreslerová, D. et al. Seeking the meaning of a unique mountain site through a multidisciplinary approach. The Late La Tène site at Sklářské Valley, Šumava Mountains, Czech Republic. Quat. Int. 542, 88–108 (2020).
Drieu, L. et al. Chemical evidence for the persistence of wine production and trade in Early Medieval Islamic Sicily. Proc. Natl Acad. Sci. USA 118, e2017983118 (2021).
Drieu, L. et al. A Neolithic without dairy? Chemical evidence from the content of ceramics from the Pendimoun rock-shelter (Castellar, France, 5750–5150 BC). J. Archaeol. Sci. Rep. 35, 102682 (2021).
Dunne, J. et al. Milk of ruminants in ceramic baby bottles from prehistoric child graves. Nature 574, 246–248 (2019).
Dunne, J., Chapman, A., Blinkhorn, P. & Evershed, R. P. Reconciling organic residue analysis, faunal, archaeobotanical and historical records: diet and the medieval peasant at West Cotton, Raunds, Northamptonshire. J. Archaeol. Sci. 107, 58–70 (2019).
Dunne, J., Chapman, A., Blinkhorn, P. & Evershed, R. P. Fit for purpose? Organic residue analysis and vessel specialisation: the perfectly utilitarian medieval pottery assemblage from West Cotton, Raunds. J. Archaeol. Sci. 120, 105178 (2020).
Dunne, J. et al. Finding Oxford’s medieval Jewry using organic residue analysis, faunal records and historical documents. Archaeol. Anthropol. Sci. 13, 48 (2021).
Evershed, R. P., Copley, M. S., Dickson, L. & Hansel, F. A. Experimental evidence for the processing of marine animal products and other commodities containing polyunsaturated fatty acids in pottery vessels. Archaeometry 50, 101–113 (2008).
Fanti, L. et al. The role of pottery in Middle Neolithic societies of western Mediterranean (Sardinia, Italy, 4500–4000 cal bc) revealed through an integrated morphometric, use-wear, biomolecular and isotopic approach. J. Archaeol. Sci. 93, 110–128 (2018).
Francés-Negro, M. et al. Neolithic to Bronze Age economy and animal management revealed using analyses of lipid residues of pottery vessels and faunal remains at El Portalón de Cueva Mayor (Sierra de Atapuerca, Spain). J. Archaeol. Sci. 131, 105380 (2021).
Gregg, M. W., Banning, E. B., Gibbs, K. & Slater, G. F. Subsistence practices and pottery use in Neolithic Jordan: molecular and isotopic evidence. J. Archaeol. Sci. 36, 937–946 (2009).
Gunnarssone, A., Oras, E., Talbot, H. M., Ilves, K. & Legzdiņa, D. Cooking for the living and the dead: lipid analyses of Rauši settlement and cemetery pottery from the 11th–13th century. Estonian J. Archaeol. 24, 45–69 (2020).
Heron, C. et al. Cooking fish and drinking milk? Patterns in pottery use in the southeastern Baltic, 3300–2400 cal bc. J. Archaeol. Sci. 63, 33–43 (2015).
Hoekman-Sites, H. A. & Giblin, J. I. Prehistoric animal use on the Great Hungarian Plain: a synthesis of isotope and residue analyses from the Neolithic and Copper Age. J. Anthropol. Archaeol. 31, 515–527 (2012).
Isaksson, S. & Hallgren, F. Lipid residue analyses of Early Neolithic funnel-beaker pottery from Skogsmossen, eastern Central Sweden, and the earliest evidence of dairying in Sweden. J. Archaeol. Sci. 39, 3600–3609 (2012).
Krueger, M., Bajčev, O., Whelton, H. L. & Evershed, R. P. in The Neolithic in the Middle Morava Valley (ed. Perić, S.) Vol. 3, 61–76 (Institute of Archaeology, 2019).
Manzano, E. et al. An integrated multianalytical approach to the reconstruction of daily activities at the Bronze Age settlement in Peñalosa (Jaén, Spain). Microchem. J. 122, 127–136 (2015).
Manzano, E. et al. Molecular and isotopic analyses on prehistoric pottery from the Virués-Martínez cave (Granada, Spain). J. Archaeol. Sci. Rep. 27, 101929 (2019).
Matlova, V. et al. Defining pottery use and animal management at the Neolithic site of Bylany (Czech Republic). J. Archaeol. Sci. Rep. 14, 262–274 (2017).
McClure, S. B. et al. Fatty acid specific δ13C values reveal earliest Mediterranean cheese production 7,200 years ago. PLoS ONE 13, e0202807 (2018).
Mileto, S., Kaiser, E., Rassamakin, Y., Whelton, H. & Evershed, R. P. Differing modes of animal exploitation in North-Pontic Eneolithic and Bronze Age Societies. Sci. Technol. Archaeol. Res. 3, 112–125 (2017).
Mukherjee, A. J., Berstan, R., Copley, M. S., Gibson, A. M. & Evershed, R. P. Compound-specific stable carbon isotopic detection of pig product processing in British Late Neolithic pottery. Antiquity 83, 743–754 (2007).
Mukherjee, A. J., Gibson, A. M. & Evershed, R. P. Trends in pig product processing at British Neolithic Grooved Ware sites traced through organic residues in potsherds. J. Archaeol. Sci. 35, 2059–2073 (2008).
Ogrinc, N., Budja, M., Potočnik, D., Žibrat Gašparič, A. & Mlekuž, D. Lipids, pots and food processing at Hočevarica, Ljubljansko barje, Slovenia. Doc. Praehist. XLI, 181–194 (2014).
Oras, E. et al. The adoption of pottery by north-east European hunter-gatherers: evidence from lipid residue analysis. J. Archaeol. Sci. 78, 112–119 (2017).
Oras, E. et al. Social food here and hereafter: multiproxy analysis of gender-specific food consumption in conversion period inhumation cemetery at Kukruse, NE-Estonia. J. Archaeol. Sci. 97, 90–101 (2018).
Outram, A. K. et al. Horses for the dead: funerary foodways in Bronze Age Kazakhstan. Antiquity 85, 116–128 (2010).
Outram, A. K. et al. Patterns of pastoralism in later Bronze Age Kazakhstan: new evidence from faunal and lipid residue analyses. J. Archaeol. Sci. 39, 2424–2435 (2012).
Özbal, H. et al. Neolitik Batı Anadolu ve Marmara yerleşimleri çanak çömleklerinde organik kalıntı analizleri. Arkeometri Sonuçları Toplantısı 28, 105–114 (2013).
Özbal, H. et al. Yenikapı, Aşağıpınar, Bademağacı ve Barcın Çömleklerindeorganik kalıntı analizi. Arkeometri Sonuçları Toplantısı 29, 83–90 (2013).
Pääkkönen, M., Bläuer, A., Evershed, R. P. & Asplund, H. Reconstructing food procurement and processing in early Comb ware period through organic residues in early Comb and Jäkärlä ware pottery. Fennosc. Archaeol. XXXIII, 57–75 (2016).
Pääkkönen, M., Bläuer, A., Olsen, B., Evershed, R. P. & Asplund, H. Contrasting patterns of prehistoric human diet and subsistence in northernmost Europe. Sci. Rep. 8, 1148 (2018).
Papakosta, V., Oras, E. & Isaksson, S. Early pottery use across the Baltic—a comparative lipid residue study on Ertebølle and Narva ceramics from coastal hunter-gatherer sites in southern Scandinavia, northern Germany and Estonia. J. Archaeol. Sci. Rep. 24, 142–151 (2019).
Pennetta, A., Fico, D., Lucrezia Savino, M., Larocca, F. & Egidio De Benedetto, G. Characterization of Bronze age pottery from the Grotte di Pertosa-Auletta (Italy): results from the first analysis of organic lipid residues. J. Archaeol. Sci. Rep. 31, 102308 (2020).
Piličiauskas, G. et al. The Corded Ware culture in the Eastern Baltic: new evidence on chronology, diet, beaker, bone and flint tool function. J. Archaeol. Sci. Rep. 21, 538–552 (2018).
Piličiauskas, G. et al. Fishers of the Corded Ware culture in the Eastern Baltic. Acta Archaeol. 91, 95–120 (2020).
Robinson, G. et al. Furness’s first farmers: evidence of Early Neolithic settlement and dairying in Cumbria. Proc. Prehist. Soc. 86, 165–198 (2020).
Robson, H. K. et al. Diet, cuisine and consumption practices of the first farmers in the southeastern Baltic. Archaeol. Anthropol. Sci. 11, 4011–4024 (2019).
Roffet-Salque, M. & Evershed, R. P. in Kopydłowo, stanowisko 6. Osady neolityczne z pogranicza Kujaw i Wielkopolski (eds Marciniak, A. et al.) 133–142 (Wydawnictwo Profil-Archeo, 2015).
Roffet-Salque, M., Banecki, B. & Evershed, R. P. in A Megalithic Tomb of the Globular Amphora Culture from Kierzkowo in the Pałuki Region—A Silent Witness of Ancestor Worship from the Stone Age Biskupin Archaeological Works (eds Nowaczyk, S. et al.) 251–266 (Archaeological Museum in Biskupin, 2017).
Roffet-Salque, M. et al. in Ludwinowo 7—Neolithic Settlement in Kuyavia Saved Archaeological Heritage 8 (Joanna Pyzel, J.) 301–316 (Profil-Archeo Publishing House and Archaeological Studio, University of Gdańsk Publishing House, 2019).
Salque, M. et al. New insights into the early Neolithic economy and management of animals in Southern and Central Europe revealed using lipid residue analyses of pottery vessels. Anthropozoologica 47, 45–61 (2012).
Smyth, J. & Evershed, R. P. The molecules of meals: new insight into Neolithic foodways. Proc. R. Ir. Acad. 115C, 27–46 (2015).
Šoberl, L. Pots for the Afterlife: Organic Residue Analysis of British Early Bronze Age Pottery from Funerary Contexts. PhD thesis. Univ. of Bristol (2011).
Šoberl, L., Žibrat Gašparič, A., Budja, M. & Evershed, R. P. Early herding practices revealed through organic residue analysis of pottery from the early Neolithic rock shelter of Mala Triglavca, Slovenia. Doc. Praehist. 35, 253–260 (2008).
Šoberl, L. et al. Neolithic and Eneolithic activities inferred from organic residue analysis of pottery from Mala Triglavca, Moverna vas and Ajdovska jama, Slovenia. Doc. Praehist. XLI, 149–179 (2014).
Spangenberg, J. E., Jacomet, S. & Schibler, J. Chemical analyses of organic residues in archaeological pottery from Arbon Bleiche 3, Switzerland—evidence for dairying in the late Neolithic. J. Archaeol. Sci. 33, 1–13 (2006).
Spangenberg, J. E., Matuschik, I., Jacomet, S. & Schibler, J. Direct evidence for the existence of dairying farms in prehistoric Central Europe (4th mil. BC). Isotopes Environ. Health Stud. 44, 189–200 (2008).
Spataro, M. et al. Production and function of Neolithic black-painted pottery from Schela Cladovei (Iron Gates, Romania). Archaeol. Anthropol. Sci. 11, 6287–6304 (2019).
Steele, V. J. & Stern, B. Red Lustrous Wheelmade ware: analysis of organic residues in Late Bronze Age trade and storage vessels from the eastern Mediterranean. J. Archaeol. Sci. Rep. 16, 641–657 (2017).
Stojanovski, D. et al. Living off the land: terrestrial-based diet and dairying in the farming communities of the Neolithic Balkans. PLoS ONE 15, e0237608 (2020).
Stojanovski, D. et al. Anta 1 de Val da Laje—the first direct view at diet, dairying practice and socio-economic aspects of pottery use in the final Neolithic of central Portugal. Quat. Int. 542, 1–8 (2020).
Tarifa-Mateo, N. et al. New insights from Neolithic pottery analyses reveal subsistence practices and pottery use in early farmers from Cueva de El Toro (Málaga, Spain). Archaeol. Anthropol. Sci. 11, 5199–5211 (2019).
Weber, J., Brozio, J. P., Müller, J. & Schwark, L. Grave gifts manifest the ritual status of cattle in Neolithic societies of northern Germany. J. Archaeol. Sci. 117, 105122 (2020).
Evershed, R. P., Heron, C. & Goad, L. J. Analysis of organic residues of archaeological origin by high-temperature gas chromatography and gas chromatography-mass spectrometry. Analyst 115, 1339–1342 (1990).
Correa-Ascencio, M. & Evershed, R. P. High throughput screening of organic residues in archaeological potsherds using direct methanolic acid extraction. Anal. Methods 6, 1330–1340 (2014).
Reimer, P. J. et al. IntCal13 and Marine13 radiocarbon age calibration curves 0–50,000 years cal bp. Radiocarbon 55, 1869–1887 (2013).
Bronk Ramsey, C. Bayesian analysis of radiocarbon dates. Radiocarbon 51, 337–360 (2009).
Graffelman, J. Exploring diallelic genetic markers: the HardyWeinberg package. J. Stat. Softw. 64, 1–23 (2015).
Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020).
Mitchell, R., Hemani, G., Dudding, T. & Paternoster, L. UK Biobank Genetic Data: MRC-IEU Quality Control Version 2; https://doi.org/10.5523/bris.1ovaau5sxunp2cv8rcy88688v (Univ. Bristol, 2019).
Brumpton, B. et al. Avoiding dynastic, assortative mating, and population stratification biases in Mendelian randomization through within-family analyses. Nat. Commun. 11, 3519 (2020).
Howe, L. J. et al. Within-sibship genome-wide association analyses decrease bias in estimates of direct genetic effects. Nat. Genet. 54, 581–592 (2022).
Agranat-Tamir, L. et al. The genomic history of the Bronze Age Southern Levant. Cell 181, 1146–1157 (2020).
Antonio, M. L. et al. Ancient Rome: a genetic crossroads of Europe and the Mediterranean. Science 366, 708–714 (2019).
Broushaki, F. et al. Early Neolithic genomes from the eastern Fertile Crescent. Science 353, 499–503 (2016).
Brunel, S. et al. Ancient genomes from present-day France unveil 7,000 years of its demographic history. Proc. Natl Acad. Sci. USA 117, 12791 (2020).
Cassidy, L. M. et al. Neolithic and Bronze Age migration to Ireland and establishment of the insular Atlantic genome. Proc. Natl Acad. Sci. USA 113, 368–373 (2015).
Cassidy, L. M. et al. A dynastic elite in monumental Neolithic society. Nature 582, 384–388 (2020).
Damgaard, Pd. B. et al. 137 ancient human genomes from across the Eurasian steppes. Nature 557, 369–374 (2018).
Damgaard, Pd. B. et al. The first horse herders and the impact of early Bronze Age steppe expansions into Asia. Science 360, eaar7711 (2018).
Feldman, M. et al. Ancient DNA sheds light on the genetic origins of early Iron Age Philistines. Sci. Adv. 5, eaax0061 (2019).
Fernandes, D. M. et al. The spread of steppe and Iranian-related ancestry in the islands of the western Mediterranean. Nat. Ecol. Evol. 4, 334–345 (2020).
Fu, Q. et al. The genetic history of Ice Age Europe. Nature 534, 200–205 (2016).
González-Fortes, G. et al. Paleogenomic evidence for multi-generational mixing between Neolithic farmers and mesolithic hunter-gatherers in the Lower Danube Basin. Curr. Biol. 27, 1801–1810 (2017).
González-Fortes, G. et al. A western route of prehistoric human migration from Africa into the Iberian Peninsula. Proc. R. Soc. B 286, 20182288 (2019).
Günther, T. et al. Population genomics of Mesolithic Scandinavia: investigating early postglacial migration routes and high-latitude adaptation. PLoS Biol. 16, e2003703 (2018).
Haber, M. et al. Continuity and admixture in the last five millennia of Levantine history from Ancient Canaanite and present-day Lebanese genome sequences. Am. J. Hum. Genet. 101, 274–282 (2017).
Harney, E. et al. Ancient DNA from Chalcolithic Israel reveals the role of population mixture in cultural transformation. Nat. Commun. 9, 3336 (2018).
Hofmanová, Z. et al. Early farmers from across Europe directly descended from Neolithic Aegeans. Proc. Natl Acad. Sci. USA 113, 6886–6891 (2016).
Järve, M. et al. Shifts in the genetic landscape of the Western Eurasian Steppe associated with the beginning and end of the Scythian dominance. Curr. Biol. 29, 2430–2441 (2019).
Jones, E. R. et al. The Neolithic transition in the Baltic was not driven by admixture with early European farmers. Curr. Biol. 27, 576–582 (2017).
Keller, A. et al. New insights into the Tyrolean Iceman's origin and phenotype as inferred by whole-genome sequencing. Nat. Commun. 3, 698 (2012).
Krzewińska, M. et al. Ancient genomes suggest the eastern Pontic-Caspian steppe as the source of western Iron Age nomads. Sci. Adv. 4, eaat4457 (2018).
Lamnidis, T. C. et al. Ancient Fennoscandian genomes reveal origin and spread of Siberian ancestry in Europe. Nat. Commun. 9, 5018 (2018).
Lazaridis, I. et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513, 409–413 (2014).
Lazaridis, I. et al. Genomic insights into the origin of farming in the ancient Near East. Nature 536, 419–424 (2016).
Lazaridis, I. et al. Genetic origins of the Minoans and Mycenaeans. Nature 548, 214–218 (2017).
Linderholm, A. et al. Corded Ware cultural complexity uncovered using genomic and isotopic analysis from south-eastern Poland. Sci. Rep. 10, 6885 (2020).
Lipson, M. et al. Parallel palaeogenomic transects reveal complex genetic history of early European farmers. Nature 551, 368–372 (2017).
Malmström, H. et al. The genomic ancestry of the Scandinavian Battle Axe Culture people and their relation to the broader Corded Ware horizon. Proc. R. Soc. B 286, 20191528 (2019).
Marcus, J. H. et al. Genetic history from the Middle Neolithic to present on the Mediterranean island of Sardinia. Nat. Commun. 11, 939 (2020).
Margaryan, A. et al. Population genomics of the Viking world. Nature 585, 390–396 (2020).
Martiniano, R. et al. The population genomics of archaeological transition in west Iberia: investigation of ancient substructure using imputation and haplotype-based methods. PLoS Genet. 13, e1006852 (2017).
Mathieson, I. et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature 528, 499–503 (2015).
Mathieson, I. et al. The genomic history of southeastern Europe. Nature 555, 197–203 (2018).
Mittnik, A. et al. Kinship-based social inequality in Bronze Age Europe. Science 93, eaax6219 (2019).
Narasimhan, V. M. et al. The formation of human populations in South and Central Asia. Science 365, eaat7487 (2019).
Olalde, I. et al. A common genetic origin for early farmers from Mediterranean Cardial and Central European LBK cultures. Mol. Biol. Evol. 32, 3132–3142 (2015).
Olalde, I. et al. The Beaker phenomenon and the genomic transformation of northwest Europe. Nature 555, 190–196 (2018).
Olalde, I. et al. The genomic history of the Iberian Peninsula over the past 8000 years. Science 363, 1230–1234 (2019).
Rivollat, M. et al. Ancient genome-wide DNA from France highlights the complexity of interactions between Mesolithic hunter-gatherers and Neolithic farmers. Sci. Adv. 6, eaaz5344 (2020).
Saag, L. et al. Extensive farming in Estonia started through a sex-biased migration from the Steppe. Curr. Biol. 27, 2185–2193 (2017).
Sánchez-Quinto, F. et al. Megalithic tombs in western and northern Neolithic Europe were linked to a kindred society. Proc. Natl Acad. Sci. USA 116, 9469–9474 (2019).
Schuenemann, V. J. et al. Ancient Egyptian mummy genomes suggest an increase of Sub-Saharan African ancestry in post-Roman periods. Nat. Commun. 8, 15694 (2017).
Schroeder, H. et al. Unraveling ancestry, kinship, and violence in a Late Neolithic mass grave. Proc. Natl Acad. Sci. USA 116, 10705–10710 (2019).
Skoglund, P. et al. Genomic diversity and admixture differs for Stone-Age Scandinavian foragers and farmers. Science 344, 747–750 (2014).
Skourtanioti, E. et al. Genomic history of Neolithic to Bronze Age Anatolia, Northern Levant, and Southern Caucasus. Cell 181, 1158–1175 (2020).
Unterländer, M. et al. Ancestry and demography and descendants of Iron Age nomads of the Eurasian Steppe. Nat. Commun. 8, 14615 (2017).
Valdiosera, C. et al. Four millennia of Iberian biomolecular prehistory illustrate the impact of prehistoric migrations at the far end of Eurasia. Proc. Natl Acad. Sci. USA 115, 3428–3433 (2018).
Villalba-Mouco, V. et al. Survival of Late Pleistocene hunter-gatherer ancestry in the Iberian Peninsula. Curr. Biol. 29, 1169–1177 (2019).
Wang, C.-C. et al. Ancient human genome-wide data from a 3000-year interval in the Caucasus corresponds with eco-geographic regions. Nat. Commun. 10, 590 (2019).
Allen Ancient DNA Resource (AADR): Downloadable genotypes of present-day and ancient DNA data. Version 44.3 https://reich.hms.harvard.edu/allen-ancient-dna-resource-aadr-downloadable-genotypes-present-day-and-ancient-dna-data (David Reich Lab, 2021).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Felsenstein, J. Theoretical Evolutionary Genetics (Univ. of Washington, 2016).
Brest, J., Greiner, S., Boskovic, B., Mernik, M. & Zumer, V. Self-adapting control parameters in differential evolution: a comparative study on numerical benchmark problems. IEEE Trans. Evol. Comput. 10, 646–657 (2006).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2018).
Revell, L. J. learnPopGen: an R package for population genetic simulation and numerical analysis. Ecol. Evol. 9, 7896–7902 (2019).
Jewett, E. M., Steinrücken, M. & Song, Y. S. The effects of population size histories on estimates of selection coefficients from time-series genetic data. Mol. Biol. Evol. 33, 3002–3027 (2016).
Weninger, B., Joris, O. & Danzeglocke, U. CalPal-2007. Cologne Radiocarbon Calibration & Palaeoclimate Research Package (CalPal, 2007).
Galate, P. BANADORA, Banque de données des dates radiocarbones de Lyon pour l’Europe et le Proche-Orient http://www.archeometrie.mom.fr/banadora (Laboratoire ArAr, 2011).
Hinz, M. et al. RADON-Radiocarbon dates online 2012. Central European database of 14C dates for the Neolithic and the Early Bronze Age. J. Neolit. Archaeol. https://doi.org/10.12766/jna.2012.65 (2012).
Manning, K., Colledge, S., Crema, E. R., Shennan, S. & Timpson, A. The cultural evolution of Neolithic Europe. EUROEVOL Dataset 1: Sites, phases and radiocarbon data. J. Open Archaeol. Data 5, e2 (2016).
Burrow, S. & Williams, S. The Wales and Borders Radiocarbon Database (Amgueddfa Cymru: National Museum Wales, 2008).
Ralston, I. & Ashmore, P. Canmore Scottish Radiocarbon Database https://canmore.org.uk/project/919374 (Historic Environment Scotland; accessed 14 July 2021).
Balsera, V., Díaz-del-Río, P., Gilman, A., Uriarte, A. & Vicent, J. M. Approaching the demography of late prehistoric Iberia through summed calibrated date probability distributions (7000–2000 cal bc). Quat. Int. 386, 208–211 (2015).
Vermeersch, P. M. Radiocarbon Palaeolithic Europe Database v.18 (KU Leuven, 2015); https://ees.kuleuven.be/geography/projects/14c-palaeolithic/index.html
Bivand, R. & Lewin-Koh, N. maptools: Tools for handling spatial objects. R package version 1.1–2 (2021).
Brownrigg, R. mapdata: Extra map databases. R package version 2.3.0 (2018).
Hijmans, R. J. raster: Geographic data analysis and modeling. R package version 3.5–15 (2022).
Bivand, R. & Rundel, C. rgeos: Interface to geometry engine—open source ('GEOS'). R package version 0.5–9 (2021).
Neuwirth, E. RColorBrewer: ColorBrewer palettes. R package version 1.1–2 (2014).
Acknowledgements
This study was funded by the European Research Council (ERC) Advanced Grant ‘NeoMilk’ FP7-IDEAS-ERC/324202. M.R.-S. thanks the Royal Society for funding her Dorothy Hodgkin Fellowship (DHF\R1\180064 and RGF\EA\181067). The Natural Environment Research Council (NERC) are thanked for partial funding of the National Environmental Isotope Facility (NEIF; NE/V003917/1). We wish to thank the NERC (NE/V003917/1), the ERC (FP7-IDEAS-ERC/340923) and the University of Bristol for funding GC–MS and GC–IRMS capabilities used for this work. Y.D. and M.G.T. received funding from the ERC Horizon 2020 research and innovation programme (grant agreement no. 788616 YMPACT) and A.T. and M.G.T. received funding from the ERC Horizon 2020 research and innovation programme (grant agreement no. 951385 COREX). G.D.S. and M.S.L. work in the MRC Integrative Epidemiology Unit at the University of Bristol (MC_UU_00011/1). D. Altoft, B. Banecki, L. Benson, P. Bickle (University of York, UK), S. Ferrandin, A. Lafarge, C. Maule (University of Bristol, UK), D. Miernecka, C. Walton-Doyle (University of Manchester, UK) and I. Wiltshire (University of Bristol, UK) are acknowledged for the sampling and/or analysis of some potsherds from this study at the University of Bristol. We thank S. Kalieva and V. Logvin (Kostanay State University, Kazakhstan), C. Lohr (Leibniz Research Institute for Archaeology, Mainz, Germany), J. Lüning (Johann Wolfgang Goethe-Universität, Frankfurt, Germany), I. Pavlů (Institute of Archaeology of the Academy of Sciences of the Czech Republic) and R. W. Schmitz (LVR-LandesMuseum, Bonn, Germany) for providing some of the sherds presented in this study. We are grateful to K. Dwyer, teaching fellow in English grammar and research methodology at University College London (UCL), for clarifying lactase non-persistence as the correct usage over non-lactase persistence, on the basis that ‘non’ qualifies persistence, even if lactase persistence is considered a compound noun. We are also grateful to L. Howe, Senior Research Associate at the MRC IEU for providing derived spousal pairs in UK Biobank. We acknowledge the use of the UCL Computer Science ECON High-Performance Computing (HPC) Cluster (ECON@UCL) and associated support services, in the completion of this work. This study was also supported by the NIHR Biomedical Research Centre at University Hospitals Bristol and Weston NHS Foundation Trust and the University of Bristol. The views expressed are those of the author(s) and not necessarily those of the NIHR or the Department of Health and Social Care.
Author information
Authors and Affiliations
Contributions
R.P.E., M.G.T. and G.D.S. conceived the overall study. M.R.-S. and R.P.E. generated new lipid residue data. M.R.-S., A.T., Y.D. and M.S.L., acquired data, assembled new databases and undertook statistical modelling. G.D.S. and M.S.L. performed the UK Biobank analyses. Y.D., A.T. and M.G.T. conceptualized the selection model likelihood analysis. Y.D. and A.T. performed the selection model testing. A.T. devised the kernel interpolation and generated Figs. 1, 2 and 3. M.G.T., R.P.E., G.D.S., M.R.-S., Y.D., A.T. and M.S.L. wrote the paper. All other authors contributed either critical archaeological information, pottery from excavations, data of various types and expert knowledge. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Daniel Wegmann, Nicola Pirastu, Shevan Wilkin and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note published maps and institutional affiliations.
Extended data figures and tables
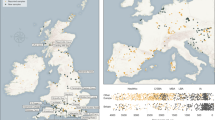
Extended Data Fig. 1 Regional fluctuations in milk use throughout European prehistory.
Percentage of milk fats through time, calculated using all animal fat residues. Grey bars and black lines illustrate 95%, 50% CI and MAP in each time slice, using a uniform prior.
Extended Data Fig. 2 Summary of model selection results for the tested ecological time series.
Inverse solar insolation, fluctuations in population level, and residential density yield models significantly better than a null model of constant selection (significance computed by likelihood ratio test). See Extended Data Table 1 for corresponding parameter estimates, and multiple testing correction (no change in the set of significant models). Abbreviations: assimilation (assi.), inverse (inv.), fluctuation (fluc.).
Extended Data Fig. 3 Inverse insolation as a driver of selection strength.
Optimized parameters, resulting selection strength- and LP allele frequency curves for inverse (inv.) insolation, one of the four ecological proxy variables yielding likelihoods significantly better than a constant selection model. Although LP is generally thought of as a dominant trait, we only show the additive model results as the parameter estimates barely differ.
Extended Data Fig. 4 Population fluctuation as a driver of selection strength.
Optimized parameters, resulting selection strength- and LP allele frequency curves for population (pop.) fluctuations (fluc.), one of the four ecological proxy variables yielding likelihoods significantly better than a constant selection model. Although LP is generally thought of as a dominant trait, we only show the additive model results as the parameter estimates barely differ.
Extended Data Fig. 5 Settlement density as a driver of selection strength.
Optimized parameters, resulting selection strength- and LP allele frequency curves for the cluster statistic, one of the four ecological proxy variables yielding likelihoods significantly better than a constant selection model. Although LP is generally thought of as a dominant trait, we only show the additive model results as the parameter estimates barely differ.
Extended Data Fig. 6 Wild animal consumption as a driver of selection strength.
Optimized parameters, resulting selection strength- and LP allele frequency curves for proportion of wild versus domestic animal, one of the four ecological proxy variables yielding likelihoods significantly better than a constant selection model. Although LP is generally thought of as a dominant trait, we only show the additive model results as the parameter estimates barely differ.
Supplementary information
Supplementary Information
Supplementary Table 1 and Figs. 1–9.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Evershed, R.P., Davey Smith, G., Roffet-Salque, M. et al. Dairying, diseases and the evolution of lactase persistence in Europe. Nature 608, 336–345 (2022). https://doi.org/10.1038/s41586-022-05010-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-05010-7
This article is cited by
-
The selection landscape and genetic legacy of ancient Eurasians
Nature (2024)
-
Unravelling the resilience of the KGK VI population from the Gumelnița site (Romania) through stable isotopes
Scientific Reports (2023)
-
Early Dalmatian farmers specialized in sheep husbandry
Scientific Reports (2023)
-
‘A Complex Past’: Theory and Applications
Journal of Archaeological Method and Theory (2023)
-
Genetic and cultural adaptations underlie the establishment of dairy pastoralism in the Tibetan Plateau
BMC Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.