Abstract
The divergence of the common dendritic cell progenitor1,2,3 (CDP) into the conventional type 1 and type 2 dendritic cell (cDC1 and cDC2, respectively) lineages4,5 is poorly understood. Some transcription factors act in the commitment of already specified progenitors—such as BATF3, which stabilizes Irf8 autoactivation at the +32 kb Irf8 enhancer4,6—but the mechanisms controlling the initial divergence of CDPs remain unknown. Here we report the transcriptional basis of CDP divergence and describe the first requirements for pre-cDC2 specification. Genetic epistasis analysis7 suggested that Nfil3 acts upstream of Id2, Batf3 and Zeb2 in cDC1 development but did not reveal its mechanism or targets. Analysis of newly generated NFIL3 reporter mice showed extremely transient NFIL3 expression during cDC1 specification. CUT&RUN and chromatin immunoprecipitation followed by sequencing identified endogenous NFIL3 binding in the –165 kb Zeb2 enhancer8 at three sites that also bind the CCAAT-enhancer-binding proteins C/EBPα and C/EBPβ. In vivo mutational analysis using CRISPR–Cas9 targeting showed that these NFIL3–C/EBP sites are functionally redundant, with C/EBPs supporting and NFIL3 repressing Zeb2 expression at these sites. A triple mutation of all three NFIL3–C/EBP sites ablated Zeb2 expression in myeloid, but not lymphoid progenitors, causing the complete loss of pre-cDC2 specification and mature cDC2 development in vivo. These mice did not generate T helper 2 (TH2) cell responses against Heligmosomoides polygyrus infection, consistent with cDC2 supporting TH2 responses to helminths9,10,11. Thus, CDP divergence into cDC1 or cDC2 is controlled by competition between NFIL3 and C/EBPs at the –165 kb Zeb2 enhancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The CUT&RUN, ChIP–seq and RNA-seq data generated in the current study are available in the Gene Expression Omnibus database with the accession number GSE188579. ChIP–seq datasets for NFIL3 in T cells were provided by V. K. Kuchroo28. ChIP–seq datasets for C/EBPβ in Ly-6Clow monocytes34 (GSE80031) and gene expression microarray datasets for Cebpa KO hematopoietic stem and progenitor cells22 (GSE146288) were downloaded and reanalyzed. Source data are provided with this paper.
References
Naik, S. H. et al. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat. Immunol. 8, 1217–1226 (2007).
Onai, N. et al. Identification of clonogenic common Flt3+ M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat. Immunol. 8, 1207–1216 (2007).
Liu, K. et al. In vivo analysis of dendritic cell development and homeostasis. Science 324, 392–397 (2009).
Grajales-Reyes, G. E. et al. Batf3 maintains autoactivation of Irf8 for commitment of a CD8alpha(+) conventional DC clonogenic progenitor. Nat. Immunol. 16, 708–717 (2015).
Schlitzer, A. et al. Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat. Immunol. 16, 718–728 (2015).
Durai, V. et al. Cryptic activation of an Irf8 enhancer governs cDC1 fate specification. Nat. Immunol. 20, 1161–1173 (2019).
Bagadia, P. et al. An Nfil3-Zeb2-Id2 pathway imposes Irf8 enhancer switching during cDC1 development. Nat. Immunol. 20, 1174–1185 (2019).
Huang, X. et al. Differential usage of transcriptional repressor Zeb2 enhancers distinguishes adult and embryonic hematopoiesis. Immunity 54, 1417–1432 (2021).
Kumamoto, Y. et al. CD301b+ dermal dendritic cells drive T helper 2 cell-mediated immunity. Immunity 39, 733–743 (2013).
Gao, Y. et al. Control of T helper 2 responses by transcription factor IRF4-dependent dendritic cells. Immunity 39, 722–732 (2013).
Tussiwand, R. et al. Klf4 expression in conventional dendritic cells is required for T helper 2 cell responses. Immunity 42, 916–928 (2015).
Murphy, T. L. et al. Transcriptional control of dendritic cell development. Annu. Rev. Immunol. 34, 93–119 (2016).
Cisse, B. et al. Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell 135, 37–48 (2008).
Scott, C. L. et al. The transcription factor Zeb2 regulates development of conventional and plasmacytoid DCs by repressing Id2. J. Exp. Med. 213, 897–911 (2016).
Wu, X. et al. Transcription factor Zeb2 regulates commitment to plasmacytoid dendritic cell and monocyte fate. Proc. Natl Acad. Sci. USA 113, 14775–14780 (2016).
Tamura, T. et al. IFN regulatory factor-4 and -8 govern dendritic cell subset development and their functional diversity. J. Immunol. 174, 2573–2581 (2005).
Bajana, S., Roach, K., Turner, S., Paul, J. & Kovats, S. IRF4 promotes cutaneous dendritic cell migration to lymph nodes during homeostasis and inflammation. J. Immunol. 189, 3368–3377 (2012).
Bajana, S., Turner, S., Paul, J., Ainsua-Enrich, E. & Kovats, S. IRF4 and IRF8 Act in CD11c+ cells to regulate terminal differentiation of lung tissue dendritic cells. J Immunol 196, 1666–1677 (2016).
Fogg, D. K. et al. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science 311, 83–87 (2006).
Hettinger, J. et al. Origin of monocytes and macrophages in a committed progenitor. Nat. Immunol. 14, 821–830 (2013).
Welner, R. S. et al. C/EBPα is required for development of dendritic cell progenitors. Blood 121, 4073–4081 (2013).
Anirudh, S. et al. TNFα rescues dendritic cell development in hematopoietic stem and progenitor cells lacking C/EBPα. Cells 9, 1223 (2020).
Zhang, W. et al. Molecular cloning and characterization of NF-IL3A, a transcriptional activator of the human interleukin-3 promoter. Mol. Cell. Biol. 15, 6055–6063 (1995).
Cowell, I. G., Skinner, A. & Hurst, H. C. Transcriptional repression by a novel member of the bZIP family of transcription factors. Mol. Cell. Biol. 12, 3070–3077 (1992).
Gilbert, L. A. et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159, 647–661 (2014).
Cress, W. D. & Triezenberg, S. J. Critical structural elements of the VP16 transcriptional activation domain. Science 251, 87–90 (1991).
Wang, G. G. et al. Quantitative production of macrophages or neutrophils ex vivo using conditional Hoxb8. Nat. Methods 3, 287–293 (2006).
Zhu, C. et al. An IL-27/NFIL3 signalling axis drives Tim-3 and IL-10 expression and T-cell dysfunction. Nat. Commun. 6, 6072 (2015).
Grant, C. E., Bailey, T. L. & Noble, W. S. FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011).
Haas, N. B., Cantwell, C. A., Johnson, P. F. & Burch, J. B. DNA-binding specificity of the PAR basic leucine zipper protein VBP partially overlaps those of the C/EBP and CREB/ATF families and is influenced by domains that flank the core basic region. Mol. Cell. Biol. 15, 1923–1932 (1995).
Ozkurt, I. C. & Tetradis, S. Parathyroid hormone-induced E4BP4/NFIL3 down-regulates transcription in osteoblasts. J. Biol. Chem. 278, 26803–26809 (2003).
Li, F., Liu, J., Jo, M. & Curry, T. E. Jr. A role for nuclear factor interleukin-3 (NFIL3), a critical transcriptional repressor, in down-regulation of periovulatory gene expression. Mol. Endocrinol. 25, 445–459 (2011).
Tamura, A. et al. C/EBPβ is required for survival of Ly6C− monocytes. Blood 130, 1809–1818 (2017).
Thomas, G. D. et al. Deleting an Nr4a1 super-enhancer subdomain ablates Ly6Clow monocytes while preserving macrophage gene function. Immunity 45, 975–987 (2016).
Heng, T. S. et al. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008).
Sathe, P., Vremec, D., Wu, L., Corcoran, L. & Shortman, K. Convergent differentiation: myeloid and lymphoid pathways to murine plasmacytoid dendritic cells. Blood 121, 11–19 (2013).
Rodrigues, P. F. et al. Distinct progenitor lineages contribute to the heterogeneity of plasmacytoid dendritic cells. Nat. Immunol. 19, 711–722 (2018).
Dress, R. J. et al. Plasmacytoid dendritic cells develop from Ly6D+ lymphoid progenitors distinct from the myeloid lineage. Nat. Immunol. 20, 852–864 (2019).
Satpathy, A. T. et al. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat. Immunol. 14, 937–948 (2013).
Anderson, D. A. III, Dutertre, C. A., Ginhoux, F. & Murphy, K. M. Genetic models of human and mouse dendritic cell development and function. Nat. Rev. Immunol. 21, 101–115 (2020).
Durai, V. & Murphy, K. M. Functions of murine dendritic cells. Immunity 45, 719–736 (2016).
Cervantes-Barragan, L. et al. Plasmacytoid dendritic cells control T-cell response to chronic viral infection. Proc. Natl Acad. Sci. USA 109, 3012–3017 (2012).
Hildner, K. et al. Batf3 deficiency reveals a critical role for CD8α+ dendritic cells in cytotoxic T cell immunity. Science 322, 1097–1100 (2008).
Shortman, K. How does batf3 determine dendritic cell development? Immunol Cell Biol 93, 681–682 (2015).
Satpathy, A. T. et al. Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J. Exp. Med. 209, 1135–1152 (2012).
Motomura, Y. et al. The transcription factor E4BP4 regulates the production of IL-10 and IL-13 in CD4+ T cells. Nat. Immunol. 12, 450–459 (2011).
Sterneck, E., Zhu, S., Ramirez, A., Jorcano, J. L. & Smart, R. C. Conditional ablation of C/EBPβ demonstrates its keratinocyte-specific requirement for cell survival and mouse skin tumorigenesis. Oncogene 25, 1272–1276 (2006).
Kamizono, S. et al. Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. J. Exp. Med. 206, 2977–2986 (2009).
Camberis, M., Le Gros, G. & Urban, J.Jr Animal model of Nippostrongylus brasiliensis and Heligmosomoides polygyrus. Curr. Protoc. Immunol. 19, 12 (2003).
Kim, S. et al. High amount of transcription factor IRF8 engages AP1–IRF composite elements in enhancers to direct type 1 conventional dendritic cell identity. Immunity 53, 759–774.e9 (2020).
Skene, P. J., Henikoff, J. G. & Henikoff, S. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat. Protoc. 13, 1006–1019 (2018).
Zhu, Q., Liu, N., Orkin, S. H. & Yuan, G. C. CUT&RUNTools: a flexible pipeline for CUT&RUN processing and footprint analysis. Genome Biol. 20, 192 (2019).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Iwata, A. et al. Quality of TCR signaling determined by differential affinities of enhancers for the composite BATF–IRF4 transcription factor complex. Nat. Immunol. 18, 563–572 (2017).
Acknowledgements
This work is supported by the US National Institutes of Health (R01AI150297, R01CA248919); D.-H.K. is supported by the National Research Foundation of Korea (NRF-2020R1A6A3A03037855); S.T.F. is a Cancer Research Institute Irvington Fellow supported by the Cancer Research Institute; R.W. is supported by the National Institutes of Health (5T32GM007200, F30CA247262); S.J.V.D. is supported by the National Institutes of Health (R01HL148033); M.S.D. is supported by NIH/NIDDK (R01DK122790-01A1); and M.K. is supported by AMED CREST (JP19gm1310002). We thank J. M. White at the Department of Pathology and Immunology Transgenic Mouse Core at Washington University in St Louis, and the Genetic Editing and iPS Cell (GEiC) centre at Washington University in St Louis for the generation of mouse models; the Genome Technology Access Center at the McDonnell Genome Institute at Washington University School of Medicine for help with genomic analysis; M. J. Murphy for advice on EMSA probe design; A. Look and T. Mak for providing Nfil3−/− mice; V. Durai for advice on embryonic stem cell targeting; H. W. Virgin for providing Hoxb8 construct; V. K. Kuchroo for providing NFIL3 ChIP–seq data in T cells; J. F. Urban Jr for providing H. polygyrus L3 larvae; and K. Saylor for technical assistance in breeding the Cebpbfl/fl mice. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government. This work benefitted from data assembled by the ImmGen consortium.
Author information
Authors and Affiliations
Contributions
T.-T.L., T.L.M. and K.M.M. designed the study. T.-T.L., S.K., F.O. and T.L.M. performed experiments, with advice from X.H., S.T.F. and R.W. P.D. performed experiments related to helminths infection. D.-H.K. and S.J.V.D. performed experiments related to ILC2 and tuft cell analysis; P.F.J. provided Cebpbfl/fl mice and advice. M.K. provided Nfil3fl/fl mice and advice. T.E., M.S.D. and S.J.V.D. provided assistance with experimental design. T.-T.L., T.L.M. and K.M.M. wrote the manuscript with contributions from all authors.
Corresponding author
Ethics declarations
Competing interests
M.S.D. is a consultant for Inbios, Vir Biotechnology, Senda Biosciences, and Carnival Corporation, and on the Scientific Advisory Boards of Moderna and Immunome. The Diamond laboratory has received unrelated funding support in sponsored research agreements from Moderna, Vir Biotechnology, Kaleido, and Emergent BioSolutions. K.M.M. is on the Scientific Advisory Board for Harbour Biomed.
Peer review
Peer review information
Nature thanks the anonymous reviewers for their contribution to the peer review of this work. Peer review reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Generation of Nfil3GFP reporter mice.
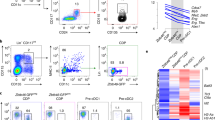
a, Schematic diagrams of mouse Nfil3 WT allele, targeting vector and targeted allele. Filled and open boxes denote coding and noncoding exons of Nfil3, respectively. N indicate NdeI site; Triangles indicate loxP sequences; TK, thymidine kinase promoter; DTA, diphtheria toxin A; pGK-Neo, neomycin selection cassette. b, Southern blot analysis of Nfil3+/+ and Nfil3GFP/+ mice. Genomic DNA was isolated from mouse liver, digested with NdeI, electrophoresed, and hybridized with Digoxigenin-labeled probes indicated in a. Southern blot with 5’ probe gave a 10.0 and a 5.0 kb band for WT and targeted allele. Southern blot with 3’ probe gave a 10.0 and a 7.7 kb band for WT and targeted allele, respectively. Progeny from ES cell clone 22 were bred to CMV-Cre mice to remove the neomycin selection cassette, and used in the following study. Data shown are one of two similar experiments. For gel source data, see Supplementary Figure 1. c, d, Frequency of cDC1 and representative flow plots showing pDCs and cDCs differentiated from WT CD117hi BM progenitors retrovirally expressing NFIL3 and GFP-NFIL3. Data are pooled from three independent experiments. e, f, Frequency of cDC1 and representative flow plots showing splenic pDCs and cDCs in WT and Nfil3GFP/GFP mice. Data are pooled from two independent experiments (n = 5 for each genotype). WT versus Nfil3GFP/GFP cDC1 P = 0.0952. g, PCA of the cDC1s and IL-4 + LPS stimulated B cells from WT, Nfil3GFP/GFP and Nfil3–/– mice by all expressed mRNAs. Mean ± s.d.; NS, not significant; f: unpaired, two-tailed Mann−Whitney test.
Extended Data Fig. 2 Nfil3GFP reporter mice analysis.
a, Representative FACS histograms of GFP-NFIL3 expression of indicated splenic cell types from WT and Nfil3GFP/GFP mice. TCRβ+ T cells were directly analyzed or stimulated with plate-bound α-CD3 + α-CD28 (2 μg/mL) antibodies for 72 h. CD19+ B cells were directly analyzed or stimulated with IL-4 (20 ng/mL) + LPS (5 μg/mL) for 24 h. Data shown are one of three similar experiments. b, Nfil3 transcripts, measured by RT-qPCR, in cells sorted from a. Data shown are one of two similar experiments. c, d, Representative flow plots showing GFP-NFIL3 expression in indicated BM cell types from WT and Nfil3GFP/GFP mice. Data shown are one of three similar experiments. For sequential gating strategies, see Supplementary Figure 1.
Extended Data Fig. 3 Nfil3 acts as a repressor to drive cDC1 specification.
a, Flow cytometric analysis of GFP-NFIL3+ BM cells from Nfil3GFP/GFP mice. Data shown are one of three similar experiments. b, c, Representative flow plots and frequencies of pDCs and cDCs differentiated from GFP-NFIL3– CDPs and GFP-NFIL3+ CDPs sorted from Nfil3GFP/GFP mice. Data are pooled from five independent experiments. d, Representative flow plots showing splenic cDC1s, cDC2s and pDCs from chimeras generated with equal mix of CD45.1+ WT BM and CD45.2+ Nfil3+/+ or Nfil3–/– BM. e, Chimerism ratio of indicated lineages in chimeras generated as in d, presented as the ratio of CD45.2+ to CD45.1+ cells normalized to the ratio of CD45.2+ to CD45.1+ LSK cells within the same mouse. Data are pooled from two independent experiments (n = 5 for Nfil3+/+ and n = 4 for Nfil3–/– mixed BM chimeras). f, g, Frequency of cDC1 and representative flow plots showing pDCs and cDCs differentiated from WT CD117hi BM progenitors retrovirally expressing NFIL3, NFIL3-KRAB or NFIL3-VP16. Data are pooled from three independent experiments. Schematic diagrams of NFIL3, NFIL3-KRAB and NFIL3-VP16 are depicted next to the plots. DBD denotes DNA-binding domain. Mean ± s.d.; e: unpaired, multiple t tests with Welch correction.
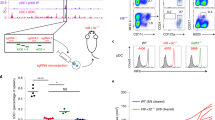
Extended Data Fig. 4 NFIL3 binds to the –165 kb Zeb2 enhancer.
a, Representative flow plots showing DC potential of Hoxb8 cell line. The Hoxb8 cells were washed twice with cold PBS to remove residual β-estradiol and cytokines before standard Flt3L culture for 7 days. b, c, Frequency of cDC1 and representative flow plots showing cDCs differentiated from Hoxb8 cells retrovirally expressing NFIL3. Data are pooled from two independent experiments (n = 6 for each cell line). d, Nfil3 transcripts, measured by RT-qPCR, in GFP-NFIL3– CDP, GFP-NFIL3+ CDP, and Hoxb8 cell lines retrovirally expressing NFIL3 used for CUT&RUN and ChIP-seq experiments. Data are pooled from three independent experiments for GFP-NFIL3– CDP, GFP-NFIL3+ CDP, and two independent experiments for Hoxb8 cell lines. e, Rank of the NFIL3 CUT&RUN and ChIP-seq peaks at the –165 kb Zeb2 enhancer. f, Alignment of human, genome draft hg19, and mouse, genome draft mm10, for the –165 kb Zeb2 enhancer regions. Red boxes indicate NFIL3 binding sites. g, EMSA showing NFIL3 binding at –165 kb Zeb2 enhancer. A 37 bp 32P-labelled DNA probe encompassing the NFIL3 binding site 1 from the –165 kb Zeb2 enhancer was synthesized as site 1 probe. A 40 bp 32P-labelled DNA probe containing an optimized NFIL3 motif and flanking sequence of the NFIL3 binding site 3 from the –165 kb Zeb2 enhancer was synthesized as N > C probe. NFIL3 binding specificity was demonstrated by competition with non-radioactively labeled probes and supershift with anti-NFIL3 antibody. The competitor 1 indicates site 1, and the competitor 1mut has the same sequence as site 1 except that the NFIL3 binding site 1 was mutated. Data shown are one of two similar experiments. For gel source data, see Supplementary Figure 1. Mean ± s.d.
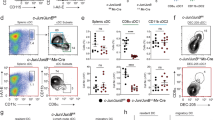
Extended Data Fig. 5 Mutation of three NFIL3 binding sites in the –165 kb Zeb2 enhancer abrogates cDC2 development (Part I).
a, Targeting strategy of NFIL3 binding site mutant mice. gRNA-1 and donor-1 were introduced into WT zygotes to mutate NFIL3 binding site 1. Δ1 mice were generated from the first round of targeting. gRNA-2, gRNA-3 and donor-2 were introduced into Δ1 homozygous zygotes to mutant NFIL3 binding site 2 and 3. Δ1+2 and Δ1+3 mice were generated from the second round of targeting. gRNA-4 and donor-3 were introduced into Δ1+2 heterozygous zygotes to mutate NFIL3 binding site 3. Δ1+2+3 mice were generated from the third round of targeting. b, Representative flow plots showing splenic cDCs and pDCs in WT, Δ1, Δ1+2, Δ1+3 and Δ1+2+3 mice. c, Number of splenic cDC1 and cDC2 in WT, Δ1, Δ1+2, Δ1+3 and Δ1+2+3 mice. Data are pooled from five independent experiments (n = 12 for WT, n = 11 for Δ1, Δ1+2, Δ1+3, and n = 8 for Δ1+2+3 mice). WT versus Δ1+2 cDC1 P = 0.2447, WT versus Δ1 cDC2 P = 0.9885, WT versus Δ1+2 cDC2 P > 0.9999. NS, not significant; c: Brown−Forsythe and Welch ANOVA with Dunnett’s T3 multiple comparisons test.
Extended Data Fig. 6 Mutation of three NFIL3 binding sites in the –165 kb Zeb2 enhancer abrogates cDC2 development (Part II).
a, Representative flow plots showing migratory and resident cDCs in mesenteric lymph nodes (MLNs) from WT and Δ1+2+3 naïve mice or mice infected with H. polygyrus (H.p.) for 14 days. b, Frequency of migratory CD24– CD172a+ cDC2 and resident CD172a+ cDC2 in MLNs from WT and Δ1+2+3 naïve mice or mice infected with H.p. for 14 days. Data are pooled from seven independent experiments (n = 9 for naïve mice and n = 5 for H.p. infected mice). c, Frequency of migratory CD103– CD11b+ cDC2 and resident CD172a+ cDC2 in SLNs from WT and Δ1+2+3 mice. Data are pooled from three independent experiments (n = 3 for each genotype). d, Frequency of CD103– CD11b+ cDC2 in small intestine lamina propria from WT and Δ1+2+3 mice. Data are pooled from four independent experiments (n = 4 for each genotype). e, Representative flow plots showing small intestine lamina propria cDCs in WT and Δ1+2+3 mice. f, Frequency and number of splenic pDC in WT, Δ1, Δ1+2, Δ1+3 and Δ1+2+3 mice. Data are pooled from five independent experiments (n = 12 for WT, n = 11 for Δ1, Δ1+2, Δ1+3, and n = 8 for Δ1+2+3 mice). g, Frequency of BM pDC in WT and Δ1+2+3 mice. Data are pooled from three independent experiments (n = 7 for each genotype). h, IFN-α, measured from splenic pDCs stimulated for 16 h with CpG-A 2216, or left unstimulated. Data are pooled from two independent experiments (n = 4 for each genotype). Mean ± s.d.; b, c, h: unpaired, multiple t tests with Welch correction; d, g: unpaired, two-tailed Mann−Whitney test; f: Brown−Forsythe and Welch ANOVA with Dunnett’s T3 multiple comparisons test.
Extended Data Fig. 7 Mutation of three NFIL3 binding sites in the –165 kb Zeb2 enhancer abrogates monocyte development.
a, Representative flow plots showing monocytes among CD45+ peripheral blood cells from WT, Δ1, Δ1+2, Δ1+3 and Δ1+2+3 mice. b, Representative flow plots showing cMoPs and monocytes (Mo) in BM of WT and Δ1+2+3 mice. Data shown are one of five similar experiments. c, Frequency of peripheral blood monocytes, splenic monocytes, and number of splenic monocytes in WT, Δ1, Δ1+2, Δ1+3 and Δ1+2+3 mice. Data are pooled from two independent experiments for peripheral blood monocytes and five independent experiments for splenic monocytes (peripheral blood monocytes: n = 13 for WT, n = 10 for Δ1, n = 8 for Δ1+2, n = 9 for Δ1+3 and n = 7 for Δ1+2+3 mice; splenic monocytes: n = 11 for WT, n = 7 for Δ1, n = 4 for Δ1+2, n = 7 for Δ1+3 and n = 8 for Δ1+2+3 mice). d, Representative flow plots showing splenic B cells and T cells in WT and Δ1+2+3 mice. e, Frequency of splenic B cells and T cells in WT, Δ1, Δ1+2, Δ1+3 and Δ1+2+3 mice. Data are pooled from three independent experiments (n = 9 for WT, n = 7 for Δ1, Δ1+2, Δ1+3, and n = 5 for Δ1+2+3 mice). WT versus Δ1+2+3 B cell P = 0.9238, WT versus Δ1+2+3 T cell P = 0.6980. f, Representative flow plots showing monocytes and macrophages in small intestine lamina propria from WT and Δ1+2+3 mice. g, Frequency of small intestine lamina propria monocytes and macrophages in WT and Δ1+2+3 mice. Data are pooled from four independent experiments (n = 4 for each genotype). Mean ± s.d.; NS, not significant; c, e: Brown−Forsythe and Welch ANOVA with Dunnett’s T3 multiple comparisons test; g: unpaired, two-tailed Mann−Whitney test.
Extended Data Fig. 8 C/EBP factors bind to the –165 kb Zeb2 enhancer to support cDC2 and monocyte development.
a, Rank of the C/EBPα and C/EBPβ CUT&RUN peaks at the –165 kb Zeb2 enhancer. b, Representative flow plots showing GFP-reporter activity in monocytes transduced with empty retrovirus (minimal CMV promoter only) or retroviruses expressing the –165 kb Zeb2 enhancer or mutants of the indicated NFIL3/C/EBP binding sites. Data shown are one of three similar experiments. A bar graph on the right shows mean values of MFI ± s.d. c, Representative flow plots showing cDCs differentiated from WT CDPs retrovirally expressing C/EBPγ or C/EBPδ. Data shown are one of two similar experiments. d, Representative flow plots showing monocytes differentiated from WT CDPs retrovirally expressing C/EBPα or C/EBPβ (LAP isoform) (pre-gate: CD317– B220– cells). cDCs expressing empty-retroviral vectors are shown as controls (dashed black lines) for Ly-6C and CD115 expression in MHCII– CD11c– cells. Data shown are one of two similar experiments. e, Frequency of cDC1s, cDC2s and monocytes in all infected cells (left panel) or infected cDCs (right panel) differentiated from WT or Δ1+2+3 CDPs retrovirally expressing C/EBPα or C/EBPβ (LAP isoform). Data are pooled from four independent experiments. f, Single-cell RNA transcriptome analysis of CDPs showing Cebpa, Cebpb and Cebpe expression visualized with Loupe Browser. g, h, Frequency of cDC2 and representative flow plots showing in vivo developmental potential of Cebpbfl/fl CD117hi BM progenitors transduced with empty retrovirus or retrovirus expressing Cre recombinase. Recipient spleens were analyzed for the presence of retroviral transduced and donor-derived cDCs. Data are pooled from two independent experiments (n = 4 for each retrovirus). i, Gene expression microarray analysis of hematopoietic stem and progenitor cells from WT or Cebpa knockout mice. The cells were left un-treated or treated with Flt3L for 4 h. Shown are averages of quadruplicate gene expression values of the indicated genes. Mean ± s.d.; h: unpaired, two-tailed Mann−Whitney test.
Extended Data Fig. 9 Myeloid and lymphoid pathways of pDC development are distinguished by different requirements in the –165 kb Zeb2 enhancer.
a, Representative flow plots showing pDCs and cDCs differentiated from sort purified CDPs and MDPs of WT and Δ1+2+3 mice, assessed after 5 days of culture with Flt3L. Data shown are one of three similar experiments. b, Representative flow plots showing in vivo developmental potential of CDPs and MDPs from WT and Δ1+2+3 mice. Recipient spleens were analyzed for the presence of CD45.2+ donor-derived pDCs and cDCs. Data shown are one of two (CDP) or three (MDP) similar experiments. c, Representative flow plots showing pDCs differentiated from sort purified CMPs and CLPs of WT and Δ1+2+3 mice, assessed after 7 (CMPs) or 5 (CLPs) days of culture with Flt3L. Data shown are one of two (CMP) or three (CLP) similar experiments. d, Representative flow plots showing CDPs (CD115+ CD127–), IL-7R+ LPs (CD115– CD127+) and IL-7R– CSF1R– NPs (CD115– CD127–) in the BM of WT and Δ1+2+3 mice (pre-gate: lineage– CD16/CD32– CD135+ CD117int-neg cells). Data shown are one of three similar experiments. e, Representative flow plots showing pDCs differentiated from sort purified CDPs, IL-7R+ LPs and IL-7R–CSF1R– NPs (as in d) of WT and Δ1+2+3 mice, assessed after 4 days of culture with Flt3L. Data shown are one of three similar experiments.
Extended Data Fig. 10 Zeb2 is not required for the maintenance of cDC2, which support TH2 responses to H. polygyrus infection.
a, Diagrams showing DC development in WT, Δ1+2+3, Irf8 +32–/– and Δ1+2+3 × Irf8 +32–/– mice. b, Representative flow plots showing monocytes among CD45+ peripheral blood cells from WT, Δ1+2+3, Irf8 +32–/– and Δ1+2+3 × Irf8 +32–/– mice. c, Frequency of peripheral blood monocytes in WT, Δ1+2+3, Irf8 +32–/– and Δ1+2+3 × Irf8 +32–/– mice. Data are pooled from two independent experiments (n = 6 for WT, Δ1+2+3, n = 7 for Irf8 +32–/– and n = 6 for Δ1+2+3 × Irf8 +32–/– mice). d, Frequency of IL-4, IL-5, IL-13 or IFN-γ expressing CD4+ T cells in MLNs from WT, Δ1+2+3, Irf8 +32–/– and Δ1+2+3 × Irf8 +32–/– naïve mice or mice infected with H.p. for 14 days (pre-gate: TCRβ+ CD4+ cells). Data are pooled from two independent experiments (n = 3 for naïve mice, n = 6 for H.p. infected WT, Δ1+2+3, Irf8 +32–/– mice and n = 5 for H.p. infected Δ1+2+3 × Irf8 +32–/– mice). WT versus Δ1+2+3 × Irf8 +32–/– IL-4 expressing CD4+ T cells in H.p. infected mice P = 0.1044, WT versus Δ1+2+3 × Irf8 +32–/– IL-13 expressing CD4+ T cells in H.p. infected mice P = 0.8243. e, f, Frequency of IgG1+ class-switched B cells (e) or FAS+ GL7+ germinal center B cells (f) of total CD19+ B cells in MLNs from WT or Δ1+2+3 naïve mice or mice infected with H.p. for 14 days (n = 3 for WT or Δ1+2+3 naïve mice, n = 5 for H.p. infected WT mice and n = 4 for H.p. infected Δ1+2+3 mice). WT versus Δ1+2+3 IgG1+ class-switched B cells in H.p. infected mice P = 0.2145. g, Total serum IgE and IgG1 in naïve WT, Δ1+2+3 mice or mice infected with H.p. for 14 days. Data are pooled from two independent experiments (n = 4 for naïve mice and n = 9 for H.p. infected mice). h, Frequency of peripheral blood eosinophils and neutrophils in WT and Δ1+2+3 mice (n = 8 for each genotype). i, Number of ILC2s per cm small intestine and frequency of ILC2s in small intestine lamina propria CD45+ cells of WT and Δ1+2+3 mice (n = 3 for each genotype). WT versus Δ1+2+3 ILC2 number P > 0.9999, frequency P = 0.4000. j, Number of tuft cells per cm small intestine and frequency of tuft cells in small intestine epithelium CD45− cells of WT and Δ1+2+3 mice (n = 3 for each genotype). WT versus Δ1+2+3 tuft cell number P = 0.7000, frequency P = 0.7000. Mean ± s.d.; NS, not significant; c: Brown−Forsythe and Welch ANOVA with Dunnett’s T3 multiple comparisons test; d: ordinary two-way ANOVA with Dunnett’s multiple comparisons test; e–g: unpaired, multiple t tests with Welch correction; h–j: unpaired, two-tailed Mann−Whitney test.
Supplementary information
Supplementary Figure 1
FACS sequential gating/sorting strategies and gel source data.
Supplementary Table 1
List of oligonucleotide primers.
Supplementary Table 2
gRNA and ssODN donor sequences for generating Δ1, Δ1+2, Δ1+3, Δ1+2+3 mice, and genomic DNA sequences of the –165 kb Zeb2 enhancer in the new mouse models.
Supplementary Table 3
List of antibodies used for flow cytometry.
Source data
Rights and permissions
About this article
Cite this article
Liu, TT., Kim, S., Desai, P. et al. Ablation of cDC2 development by triple mutations within the Zeb2 enhancer. Nature 607, 142–148 (2022). https://doi.org/10.1038/s41586-022-04866-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-04866-z
This article is cited by
-
An engineered ligand-responsive Csy4 endoribonuclease controls transgene expression from Sendai virus vectors
Journal of Biological Engineering (2024)
-
Dendritic cells as orchestrators of anticancer immunity and immunotherapy
Nature Reviews Clinical Oncology (2024)
-
Distinct ontogenetic lineages dictate cDC2 heterogeneity
Nature Immunology (2024)
-
Cell-specific and shared regulatory elements control a multigene locus active in mammary and salivary glands
Nature Communications (2023)
-
The transcription factor Zeb1 controls homeostasis and function of type 1 conventional dendritic cells
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.