Abstract
SARS-CoV-2, like other coronaviruses, builds a membrane-bound replication organelle to enable RNA replication1. The SARS-CoV-2 replication organelle is composed of double-membrane vesicles (DMVs) that are tethered to the endoplasmic reticulum (ER) by thin membrane connectors2, but the viral proteins and the host factors involved remain unknown. Here we identify the viral non-structural proteins (NSPs) that generate the SARS-CoV-2 replication organelle. NSP3 and NSP4 generate the DMVs, whereas NSP6, through oligomerization and an amphipathic helix, zippers ER membranes and establishes the connectors. The NSP6(ΔSGF) mutant, which arose independently in the Alpha, Beta, Gamma, Eta, Iota and Lambda variants of SARS-CoV-2, behaves as a gain-of-function mutant with a higher ER-zippering activity. We identified three main roles for NSP6: first, to act as a filter in communication between the replication organelle and the ER, by allowing lipid flow but restricting the access of ER luminal proteins to the DMVs; second, to position and organize DMV clusters; and third, to mediate contact with lipid droplets (LDs) through the LD-tethering complex DFCP1–RAB18. NSP6 thus acts as an organizer of DMV clusters and can provide a selective means of refurbishing them with LD-derived lipids. Notably, both properly formed NSP6 connectors and LDs are required for the replication of SARS-CoV-2. Our findings provide insight into the biological activity of NSP6 of SARS-CoV-2 and of other coronaviruses, and have the potential to fuel the search for broad antiviral agents.
Similar content being viewed by others
Main
SARS-CoV-2 extensively rearranges host cellular membranes into replication organelles that provide a microenvironment conducive to RNA synthesis and protection from host sensor and defence systems1,2. The 16 viral NSPs that are released from polyproteins pp1a and pp1ab by 2 viral proteases include 13 cytosolic proteins, which are involved in RNA replication, and 3 transmembrane proteins, NSP3, NSP4 and NSP6. Studies on other coronaviruses suggest that NSP3 and NSP4, with a hitherto undefined contribution from NSP6, are responsible for generating the replication organelles3,4,5,6. Despite considerable advances in our understanding of the ultrastructure of the SARS-CoV-2 replication organelle2,7,8, mechanistic insights into its biogenesis have so far been limited. In particular, there is at present—to our knowledge—no information on the role of NSP6 in this process. Of note, six SARS-CoV-2 ‘variants of concern’ (VOCs) (Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Eta (B.1.525), Iota (B.1.526)9 and Lambda (C.37)10) share a three-amino-acid deletion in NSP6 (NSP6(ΔSGF)), in addition to the more noted mutations in the spike protein; this finding adds further impetus to the need to examine the role of NSP6 in the biogenesis of replication organelles and in the replication of SARS-CoV-2.
NSP6 induces ER zippering
We tagged SARS-CoV-2 NSP6 at either the N or the C terminus. C-terminally tagged NSP6 showed a diffuse distribution in the ER (Fig. 1a and Extended Data Fig. 1a), as reported for NSP6 from other coronaviruses3,11. Conversely, N-terminally tagged NSP6, expressed at a comparable level, and untagged NSP6 elicited the formation of roundish structures (Fig. 1a and Extended Data Fig. 1a–c). These structures, which we refer to as the NSP6 compartment, did not colocalize with endosomal, lysosomal or autophagosomal markers (Extended Data Fig. 1d), but colocalized with the ER-reporter protein Cb5 (the C-terminal tail of cytochrome b5)12 (Fig. 1a and Extended Data Fig. 1a). This appears to be a general feature of coronavirus NSP6, as N- but not C-terminally tagged avian infectious bronchitis virus (IBV) NSP6 also formed roundish structures that colocalized with Cb5 (Extended Data Fig. 1e).
a, HeLa cells expressing YFP–Cb5 alone, co-expressing C- or N-terminally Flag-tagged NSP6, or co-expressing untagged NSP6. Insets, enlarged merged images of boxed areas; arrowheads, NSP6 compartments; dashed lines, cell boundaries. b,c, IEM (anti-haemagglutinin (HA) immunolabelling) (b) and EM (c) of HeLa cells expressing HA–NSP6. White arrowheads, linear zippered ER membranes; black arrowheads, circular zippered ER membranes; black arrows, continuity between zippered and regular ER membranes. The regular ER is shown in green. The average size of circular NSP6-positive ER structures is 623 ± 231 nm. d, Morphometric analysis of NSP6-expressing cells (percentage of the ER surface that is associated with regular cisternae or zippered domains). Mean ± s.d., N = 3, n = 60 (in all figure legends, N indicates the number of experiments and n the number of total measurements or observations). e–h, Immuno-CLEM analysis of the NSP6 compartments. e, Fluoromicrograph of HA–NSP6. f,g, Enlargement with NSP6-labelled structures 1–7 that were identified on EM serial sections (g, left), and correspondence of NSP6 fluorescent spots with NSP6 circular and linear zippered ER membranes (g, right). h, Serial sections of structure 6 in g. Black arrows, NSP6-positive linear zippered membrane connections with ER cisternae. White arrows, NSP6-positive circular zippered structures. i, HeLa cells co-expressing NSP6 with CLRT or ATL2 as indicated. Insets, enlarged merged images of boxed areas; arrowheads indicate co-localization. j,k, FRAP analysis of GFP–VAP-A and mCherry–NSP6 co-expressing cells. j, NSP6 compartments (boxed) were photobleached and the fluorescence recovery was monitored. The small panels are representative frames showing the time in seconds after the bleach (see Supplementary Video 2). k, Quantitative FRAP analysis of the reporters in j and in Extended Data Fig. 2e,f. VAP-A ER, FRAP of VAP-A in areas of regular ER; PC, BODIPY C12-HPC. Fluorescence intensity is expressed as a percentage of the value measured at time 0 (T0), which is normalized to 100%. Mean ± s.d., N = 3, n = 45. Scale bars, 10 μm (a,i,j); 120 nm (b,c); 4 µm (e); 1.1 µm (f,g); 250 nm (h).
Immunoelectron microscopy (IEM) showed that NSP6 was highly concentrated on ER cisternae that had tightly juxtaposed delimiting membranes, leaving a barely visible lumen (Fig. 1b and Extended Data Fig. 1f,g). These structures—which we refer to as zippered ER, by analogy with those reported for other viruses4,6—were highly reminiscent of the 'ER connectors' that are observed in SARS-CoV-2-infected cells2. The zippered ER structures were either linear or, more often, circular structures that encapsulated the neighbouring cytoplasm (Fig. 1c,d). Clear connections between these zippered ER structures and the regular ER were visible by electron microscopy (EM) and electron tomography (Fig. 1c, Extended Data Fig. 1h–j and Supplementary Video 1). Correlative light and electron microscopy (CLEM) showed that the roundish or elongated NSP6 spots that were visualized by immunofluorescence corresponded, respectively, to the circular or linear zippered ER profiles that were observed by EM (Fig. 1e–g), whose connection to the regular ER can be traced (Fig. 1h). The NSP6-containing structures were not freely accessible to ER luminal proteins (such as calreticulin and the ER reporter GFP-KDEL) or to ER membrane proteins with bulky luminal domains (such as ERGIC53 and ATF6) but were accessible to ER membrane proteins such as VAP-A, atlastin-2 and the KDEL receptor that possess no or very small luminal tracts (Fig. 1i–k and Extended Data Fig. 2a–d). We validated the continuity between the NSP6 compartment and the ER using fluorescence recovery after bleaching (FRAP) assays (Fig. 1j,k, Extended Data Fig. 2e and Supplementary Videos 2 and 3). After bleaching, both VAP-A and Cb5 re-entered the NSP6 compartment, although with slower kinetics compared with the regular ER. The NSP6 compartment was accessible to phospholipids, such as BODIPY C12-HPC, whose fluorescence also recovered after bleaching (Fig. 1k, Extended Data Fig. 2f and Supplementary Video 4). NSP6 itself, however, showed limited FRAP (Fig. 1j,k and Extended Data Fig. 2e,f), probably because it is engaged in stable protein complexes.
Our results indicate that NSP6 drives the formation of a zippered double-membrane compartment that maintains continuity with the ER but largely excludes ER luminal proteins.
NSP6 homodimers zipper ER membranes
The structure of NSP6 has not been solved and different topologies have been predicted. The N and C termini of NSP6 must face the cytosol because they are processed by the cytosolic NSP5 protease. Indeed, N- or C-terminally tagged NSP6 was readily detectable by antibodies after plasma membrane permeabilization (Extended Data Fig. 3a). On the basis of these data, topology predictions using the CCTOP server13 and biochemical analyses of other coronaviruses14,15, we assigned six transmembrane domains to NSP6 and envisaged that the seventh predicted transmembrane domain, which is an amphipathic helix16, does not cross, but remains associated with, the membrane (Fig. 2a and Extended Data Fig. 3b).
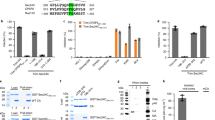
a, Predicted secondary structure of NSP6. The ΔSGF deletion and truncation site (at residue 157) are indicated. b, HeLa cells expressing Myc–NSP6 (inset) and/or Flag–NSP6(1–157). The fraction of NSP6(1–157) that is associated with NSP6 structures is indicated. Mean ± s.d., N = 3, n = 74. c, Cells expressing GFP–NSP6 alone or together with mCherry–NSP6. Bottom graph, FRET measurements in cells co-expressing mCherry–NSP6 with the indicated GFP-tagged protein. Mean ± s.d., N = 3, n = 20. d, Immunoprecipitation (IP) and western blot (WB) from GFP–NSP6 and HA–NSP6 co-expressing cells, representative of four independent experiments. e, Radial layout of a phylogenetic tree of 3,508 SARS-CoV-2 genomes. VOCs are indicated and the percentage of each genome containing ΔSGF is reported. Black branches highlight the appearance of the deletion. f, Mutations involved in the branching and specificity of each VOC. Arrows, appearance of the ΔSGF and mutations in the spike (S) protein. g,h, Time course analysis of stably expressing Flag–NSP6 or Flag–NSP6(ΔSGF) cells induced with doxycycline. g, Fluoromicrographs at 3 h. h, Quantification of the structures shown in g and Extended Data Fig. 5a (N = 3, n = 90). Left, number and areas of NSP6-positive structures. One pixel corresponds to 0.0825 μm. Right, NSP6 in structures as a percentage of the total NSP6 in the cell (mean ± s.e.m.). i,j, EM (i) and IEM (j) (anti-HA immunolabelling) of HA–NSP6(ΔSGF)-expressing HeLa cells. White arrowheads, linear zippered ER structures; black arrowheads, circular zippered ER structures. The regular ER is shown in green. k, Morphometric analysis of IEM images. Quantification of gold particles at zippered ER (percentage of the total ER-associated-particles). l, The surface area of zippered ER normalized for the total number of gold particles. For k,l, N = 3, n = 19. Scale bars, 10 µm (b,c,g); 250 nm (i,j). Two-tailed Mann–Whitney test (c); unpaired two-tailed t-test (k,l); one-way ANOVA with Tukey’s post-hoc test (h). NS, not significant.
Truncating the C-terminal part of NSP6 including the amphipathic helix (NSP6(1–157); Fig. 2a) or introducing two mutations that abrogate its amphiphilic properties16 (NSP6(F220Q/T222W); Extended Data Fig. 3b) caused NSP6 to distribute diffusely in the ER (Fig. 2b and Extended Data Fig. 3c,d). However, although necessary, the amphipathic helix is not sufficient to induce ER remodelling as the C-terminal domain—which includes part of the amphipathic helix (NSP6-C80; see below)—was unable to induce it. We found that homodimerization of NSP6 is also required. Fluorescence resonance energy transfer (FRET) measurements and the co-immunoprecipitation of GFP–NSP6 co-expressed with mCherry–NSP6 or HA–NSP6 indicated that NSP6 undergoes homodimerization (Fig. 2c,d and Extended Data Fig. 3e,f). Dimerization involves amino acids 1–157, as NSP6(1–157) was massively recruited and retained in the NSP6 compartment when co-expressed with the full length NSP6 (Fig. 2b). Indeed, both FRET and co-immunoprecipitation experiments (Fig. 2c and Extended Data Fig. 3g,h) confirmed that NSP6(1–157) and NSP6 interact, which indicates that NSP6(1–157) maintains the homodimerization interface(s).
Together, these data indicate that both the C-terminal amphipathic helix and NSP6 homodimerization (through amino acids 1–157) are required to generate the NSP6 compartment.
K22, a small molecule that interferes with the replication of several coronaviruses with different potencies, has been hypothesized to target NSP6, as K22-resistant strains of human coronavirus 229E (HCoV-229E) have mutations in NSP6 (ref. 17). We found that K22 (at 40 µM) reduced the number of regular NSP6 structures and NSP6 retention in these structures (Extended Data Fig. 4a,b). In addition, 37% of K22-treated cells exhibited elongated perinuclear NSP6 structures (Extended Data Fig. 4a,c). Immuno-CLEM (Extended Data Fig. 4d–f) and EM (Extended Data Fig. 4g–j) showed that these structures corresponded to extensive zippered areas of the nuclear envelope. Thus, impaired formation of the NSP6 compartment induced by K22 may be due in part to a shift in NSP6 zippering activity towards the nuclear envelope, which is apparently an unfavourable site for replication organelle formation (https://www.ebi.ac.uk/empiar/EMPIAR-10490/).
NSP6(ΔSGF) has a higher ER-zippering activity
Six SARS-CoV-2 VOCs (Alpha, Beta, Gamma, Eta, Iota and Lambda) have a three-amino-acid deletion (SGF, positions 106–108) in the predicted second and longest NSP6 luminal loop. Phylogenetic analysis of SARS-CoV-2 using Nextstrain18 showed that the deletion emerged independently in these lineages (Fig. 2e,f), suggesting that it conveys a selective advantage.
We found that, compared with the Wuhan-Hu-1 NSP6 (from here on, the reference NSP6), NSP6(ΔSGF) is more proficient in inducing the NSP6 compartment: the kinetics of formation were faster (Fig. 2g,h, Extended Data Fig. 5a); the NSP6(ΔSGF) compartments were more numerous and larger (Fig. 2h); and NSP6(ΔSGF) was more enriched in these compartments (Fig. 2h). These differences were not due to different protein levels or half-lives (Extended Data Fig. 5b,c), but to the higher propensity of NSP6(ΔSGF) to homo-oligomerize, as indicated by its higher resistance to detergent extraction (Extended Data Fig. 5d), more efficient co-immunoprecipitation (Extended Data Fig. 5e) and lower mobility (in fluorescence loss in photobleaching (FLIP) and FRAP experiments) compared to the reference NSP6 (Extended Data Fig. 5f–h). NSP6(ΔSGF) was slightly less sensitive than the reference NSP6 to K22 (Extended Data Fig. 5g–i). Finally, EM, IEM and CLEM showed that NSP6(ΔSGF) promoted the formation of both linear and circular zippered membrane compartments (Fig. 2i,j and Extended Data Fig. 5j–l), like NSP6, but that it was more highly associated with zippered membrane domains and was depleted from the regular ER (Fig. 2j,k). This was paralleled by an increase in the ER surface area occupied by zippered domains (Fig. 2l).
The higher ER-zippering activity of NSP6(ΔSGF) was also evident comparing the putative precursor of NSP6; that is, NSP6–NSP7 and NSP6(ΔSGF)–NSP7. During viral infection, NSP6 is generated by polyprotein cleavage by NSP5 (ref.1). Consistent with NSP6 forming the NSP6 compartment only if its C terminus is ‘free’, NSP6–NSP7 showed a diffuse ER distribution (Extended Data Fig. 5m,n) and also a partial Golgi localization, suggesting that the precursor might visit the Golgi before the cleavage unleashes its ER-zippering activity. By contrast, NSP6(ΔSGF)–NSP7 was mainly retained in the ER and was able to form small roundish structures even before cleavage (Extended Data Fig. 5m,n).
NSP6 connects DMVs with the ER
Given the similarity of NSP6-induced zippered ER to ‘ER connectors’ between the ER and DMVs in SARS-CoV-2-infected cells2, we investigated the relationship between NSP6 and the DMVs. When expressed alone, NSP3 and NSP4 exhibited a diffuse distribution in the ER (Extended Data Fig. 6a), but when co-expressed (hereafter referred to as NSP3/NSP4), and in agreement with recent reports19,20, they fully colocalized in punctate structures (Extended Data Fig. 6b,c). At the EM level, these corresponded to clusters of vesicles that had a diameter of 50–100 nm and were surrounded by two membranes (that is, DMVs), with a visible intermembrane space (Extended Data Fig. 6d,e).
Thus, NSP3/NSP4 and NSP6 are individually able to reproduce the two main features of the SARS-CoV-2 replication organelle, DMVs and the connectors2, respectively.
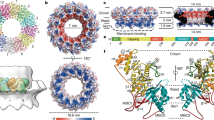
The combined expression of all three membrane NSPs (NSP3/NSP4/NSP6; Extended Data Fig. 6f,g) revealed NSP3/NSP4-positive puncta in close proximity to but not overlapping the NSP6 compartment (Fig. 3a). Notably, a similar segregation of NSP6- and NSP3-positive domains was also detectable in Calu-3 cells infected with an early lineage or the Gamma variant of SARS-CoV-2 (Fig. 3b). CLEM revealed that the NSP3/NSP4 puncta corresponded to clusters of DMVs, whereas the NSP6 structures corresponded to tracts of zippered ER that remained distinct from but were often close and connected to the DMVs (Fig. 3c,d). IEM showed that groups of NSP3/NSP4-positive DMVs associated with NSP6-positive zippered ER membranes (Fig. 3e). Tomographic analysis of NSP3/NSP4/NSP6-expressing cells revealed that the DMVs were organized in ’grape-like’ clusters, sometimes with reciprocal connections (Fig. 3f,g). Long tracts of zippered ER formed connections between the DMV clusters and the ER proper (Fig. 3f,g and Supplementary Videos 5–7), similar to those observed in SARS-CoV-2-infected cells2. Thus, we inferred that NSP6 forms the zippered connectors that guarantee full membrane—but restricted luminal—continuity with the ER.
a, Calu-3 cells expressing HA–NSP3 + Flag–NSP6 + mCherry–NSP4. The enlargement of the boxed area shows HA and Flag immunolabelling. b, Calu-3 cells infected with early lineage and Gamma variant SARS-CoV-2. Values represent mean NSP3- and NSP6-structure distances in nm. N = 10 cells, n = 2,233 structures. c,d, CLEM. Fluoromicrograph (c) and EM serial sections (d) of an HA–NSP3 + mCherry–NSP4 + GFP–NSP6–expressing cell. Arrowheads, NSP3/NSP4 co-localization; arrows, NSP6 compartments; black arrow, NSP6 compartment connection with NSP3/NSP4 DMVs. e, IEM showing NSP3 (anti-HA, white arrowheads) and NSP6 (anti-GFP, white arrows). f,g, Tomogram (f) and three-dimensional (3D) reconstruction (g) showing connections of zippered ER to DMVs (white arrow and arrowhead) and to the regular ER (black arrow) (Supplementary Videos 5 and 6). h, Number and distribution of NSP4 puncta in cells expressing the indicated NSPs. Mean ± s.d., N = 3, n = 30. Box plots represent 25th to 75th percentiles of the data (centre line, median; whiskers, minima and maxima). i,j, Tomograms (i) and 3D reconstruction (j) showing multiple short DMV–ER tubular connections (white arrows) in an NSP3/NSP4-expressing cell (Supplementary Videos 8 and 9). k, Tomograms from NSP3/NSP4 and NSP3/NSP4/NSP6-expressing cells showing DMV–ER connections (arrows). l, Intensity profiles along the red lines in k. m, Morphometry of NSP3/NSP4- or NSP3/NSP4/NSP6-expressing cells. n,o, Tomogram (n) and 3D reconstruction (o) of a NSP3/NSP4/NSP6(ΔSGF)-expressing cell, showing numerous zippered ER domains connected to DMVs (white arrows) and to regular ER (black arrows) (Supplementary Videos 11 and 12). p, Tomograms showing zippered ER connections (arrows) to DMVs in Calu-3 cells infected with early lineage and Gamma variant SARS-CoV-2 (Supplementary Videos 13 and 14). q, Length of zippered ER connected to DMVs in infected cells (N = 10, n ≥ 20), and from early lineage* (Bavpat1/2020) data in EMPIAR-10490 (29 tomograms). Scale bars, 10 µm (a,b); 10 µm (c); 470 nm (d); 200 nm (e,p); 160 nm (f,g,i,j,n,o); 100 nm (k). One-way ANOVA with Tukey’s (h, left) or Emmeans (h, right) post-hoc test; unpaired two-tailed t-test (m,q). NS, not significant.
We then asked how NSP3/NSP4-induced DMVs might be affected by NSP6. The NSP3/NSP4 puncta were more numerous and more homogeneously distributed throughout the cytoplasm in NSP3/NSP4/NSP6-expressing (both reference and NSP6(ΔSGF)) than in NSP3/NSP4-expressing cells (Fig. 3h), suggesting that NSP6 might provide a cue for the positioning and organization of DMVs (Fig. 3a,c).
Electron tomography revealed that in the absence of NSP6, DMV connections with the ER were short and tubular with a clearly detectable lumen (Fig. 3i–m, Extended Data Fig. 6h and Supplementary Videos 8–10). By contrast, in the presence of NSP6, DMV clusters were connected with the ER through much longer sheet-like zippered domains (Fig. 3f,g,k–m, Extended Data Fig. 6h and Supplementary Videos 5–7). The number of DMVs per connection was also different: an average of around 3 DMVs per tubular connection without NSP6 and of around 15 DMVs per zippered connection with NSP6 (Fig. 3m). In addition, the shape of the DMVs was more regular (Fig. 3m), their size was more uniform (Extended Data Fig. 6i–k) and their packing inside each cluster was denser in the presence of NSP6 (Fig. 3f,g,i,j and Extended Data Fig. 6l–n).
These data indicate that the co-expressed NSP3/NSP4/NSP6 reproduce replication-organelle-like structures (ROLS) and that NSP6 organizes DMV clusters.
We then assessed whether conditions that negatively or positively affect the ER-zippering activity of NSP6—that is, K22 treatment or SGF deletion, respectively—have an effect on the ROLS. Although K22 had no effect on the number and distribution of NSP3/NSP4 puncta in cells expressing only NSP3 and NSP4, it blunted the ability of co-expressed NSP6 to increase the number of NSP3/NSP4 puncta (Extended Data Fig. 7a,b). EM revealed that DMV clusters in these cells contained a significantly lower number of vesicles (Extended Data Fig. 7c–f) with a less regular shape (Extended Data Fig. 7e,f) that lost zippered connections and acquired more tubular connections to the ER (Extended Data Fig. 7d–f). Thus, treatment with K22 counteracted the ability of NSP6 to form zippered connections and to promote the homogeneous growth of DMVs. Corroborating these results, we found that K22, at the (relatively high) concentrations that interfere with the biogenesis of ROLS (40 μM), but not at lower ones21, inhibited the replication of SARS-CoV-2 (Extended Data Fig. 7g–i).
As for the SGF deletion, we found that NSP6(ΔSGF) also enhances and organizes the formation of NSP3/NSP4 puncta (Fig. 3h), but that each DMV cluster contains a higher number of DMVs that are more homogeneous in terms of size, as well as exhibiting a more developed system of zippered connections compared to the reference NSP6 (Fig. 3n,o, Extended Data Fig. 8a–e and Supplementary Videos 11 and 12).
Finally, we analysed the zippered connectors in Calu-3 cells infected with an early lineage or Gamma variant SARS-CoV-2 that contains the SGF deletion in NSP6, and found that the Gamma strain has a much more extensive zippered connector system joining the DMVs with each other and with the ER (Fig. 3p–q, Extended Data Fig. 8f–h and Supplementary Videos 13 and 14). One might speculate that the higher zippering activity of NSP6(ΔSGF) has a role in establishing a more functional and better shielded replication organelle, providing one of the multiple mechanisms that contribute to the reported differences in replication dynamics and immune evasion of NSP6(ΔSGF)-bearing VOCs22,23.
NSP6 mediates replication organelle–LD association
A C-terminal 80-amino-acid fragment of NSP6 (NSP6-C80), which is unable to induce the formation of the NSP6 compartment, was associated with roundish cytoplasmic structures. These were negative for endosomal, Golgi or mitochondrial markers, but turned out to be lipid droplets (LDs) (Extended Data Fig. 9a,b). This association is due to the amphipathic helix, as a mutated version of NSP6-C80 (NSP6-C80(F220Q/T222W)) that lost the amphiphilic properties of its amphipathic helix did not associate with LDs and exhibited a diffuse distribution (Extended Data Fig. 9c).
Of note, and in agreement with previous results24, we found that 40% of the viral replication areas labelled by double-stranded RNA (dsRNA) and NSP6 are associated with LDs (Extended Data Fig. 9d), and that LDs are required for the replication of SARS-CoV-2 in Calu-3 cells because A922500—an inhibitor of DGAT-1—inhibited LD biogenesis and significantly reduced the viral load (Extended Data Fig. 9e).
We found that LDs were also in close proximity to ROLS in cells that co-expressed NSP3/NSP4/NSP6, mimicking the situation of infected cells, but not in cells that expressed only NSP3/NSP4 (Fig. 4a,b and Extended Data Fig. 9f,g). By contrast, LDs were found very close to NSP6 structures in cells that expressed NSP6 alone (Fig. 4a,b and Extended Data Fig. 9g). These data indicate that NSP6 mediates the association of LDs with ROLS.
a, HeLa cells expressing the indicated NSPs stained with BODIPY-488 for LDs (green). Insets, enlargement of boxed area; arrowheads, LDs close to ROLS (left) or to NSP6 (right). Bottom panels, green circles delineate the position of LDs. b, Distance of LDs from NSP4 and NSP6 puncta measured in whole cells expressing the indicated NSPs (see Methods). LD to NSP4 puncta, N = 3; n = 1,692 in NSP3/NSP4-expressing and n = 2,971 in NSP3/NSP4/NSP6-expressing cells. LD to NSP6 puncta, N = 3; n = 3,239 in NSP6-expressing and n = 2,563 in NSP3/NSP4/NSP6-expressing cells. c,d, HeLa cells expressing GFP–DFCP1 alone or with mCherry–NSP4 + HA–NSP3 + Flag–NSP6 (c), or with mCherry–NSP6 (d). Arrowheads, DFCP1 signal in the NSP6 compartment. In d, the percentage of co-localization between DFCP1 and NSP6 is indicated (mean ± s.d., N = 3, n = 30). e, FLIM–FRET analysis showing the average GFP lifetime in HeLa cells expressing GFP–NSP6 with mCherry or with mCherry–DFCP1. Mean ± s.d., N = 3, n = 15. f, LD staining (BODIPY-488, green) of control (mock-transfected) and DFCP1-knockdown (DFCP1-KD) cells expressing mCherry–NSP4 + HA–NSP3, mCherry–NSP4 + HA–NSP3 + Flag–NSP6 or non-transfected (NT). Insets, mCherry–NSP4 fluorescence (red) and anti-Flag immunostaining (blue). Graph, quantification of LD area in cells. Values are normalized to the NT-cells in either the control or the DFCP1-KD condition. The significance of LD area reduction after DFCP1-KD and mock treatment was assessed as described in the Methods. N = 3, n = 90. g, Number of NSP4 puncta per cell in mCherry–NSP4 + HA–NSP3 + Flag–NSP6-transfected cells without (control) or with DFCP1-KD. N = 3; control n = 66, DFCP1-KD n = 71. Scale bars, 10 μm (a,c,d,f). Kruskal–Wallis test with Wilcoxon post-hoc and Bonferroni correction (b); unpaired two-tailed t-test (e); Wilcoxon test (f); unpaired two-tailed Mann–Whitney test (g). NS, not significant. Box plots in b,f,g represent 25th to 75th percentiles of the data (centre line, median; whiskers, minima and maxima).
We investigated the involvement of molecular complexes that are known to tether LDs to the ER25,26,27 and found that DFCP1 (Fig. 4c) and RAB18 (Extended Data Fig. 9h) were associated with ROLS. In particular, DFCP1 was recruited by NSP6 but not by NSP3/NSP4 (Fig. 4d and Extended Data Fig. 9i). We found that the two proteins interact, as shown by the intense FRET signal that was measured in cells expressing GFP–NSP6 and mCherry–DFCP1 (Fig. 4e) and by the ability of DFCP1 to pull down NSP6 from the lysates of cells expressing HA–NSP6 (Extended Data Fig. 9j). The C-terminal domain of NSP6 mediates the recruitment of DFCP1, as NSP6(1–157) was unable to recruit DFCP1 (Extended Data Fig. 10a). A DFCP1 mutant (DFCP1(Δ1–416)) that lacks the N-terminal domain but includes the ER-targeting domain and the two FYVE domains28 is still recruited by NSP6 (Extended Data Fig. 10b), as is the FYVE domain mutant C654S/C770S, which is unable to bind phosphatidylinositol 3-phosphate (PtdIns3P) (but not the single-point mutant W543A in the ER domain) (Extended Data Fig. 10b). Thus, unlike recruitment to the omegasome (the site of autophagosome formation)29, recruitment of DFCP1 to the ROLS is independent of PtdIns3P. Indeed, inhibition of PtdIns3P generation, by wortmannin or SAR405, did not affect the recruitment of DFCP1 to the NSP6 compartment (Extended Data Fig. 10c). SAR405 did not impair the formation of the NSP6 compartment, which argues against a role for PtdIns3P in this process. Supporting an autophagy-independent role of DFCP1 recruitment by NSP6, the number of autophagosomes in cells expressing NSP6 was comparable to that of non-transfected cells (Extended Data Fig. 10d).
Notably, we found that LDs are consumed during the formation of ROLS in cells expressing NSP3/NSP4/NSP6, but not in cells expressing NSP3/NSP4 (Fig. 4f), and that a fluorescent fatty acid incorporated into LDs30 shows more efficient transfer to NSP3/NSP4 structures in the presence of NSP6 (Extended Data Fig. 10e)—consistent with a role for NSP6 in channelling LD-derived lipids to the ROLS. Of note, the NSP6-dependent consumption of LDs and the formation of ROLS were both inhibited by the knockdown of DFCP1 (Fig. 4f,g and Extended Data Fig. 10f). Finally, and in line with recent reports19, depletion of DFCP1 also inhibited the replication of SARS-CoV-2, confirming that the availability of LDs is required to sustain viral replication (Extended Data Fig. 10g,h).
Conclusions
The SARS-CoV-2 replication organelle is made of DMVs and connectors2,8, the molecular determinants of which we have shown are constituted by NSP3/NSP4 and NSP6, respectively. The NSP6 connectors are tracts of zippered ER that are not accessible to luminal ER proteins or ER membrane proteins with bulky luminal domains, but which are freely accessible to lipids (Extended Data Fig. 11). In addition to linking the DMVs to the ER, the connectors mediate the association of replication organelles with LDs (Extended Data Fig. 11); this is likely to be a dynamic association (as at any given time, 40% of replication organelles are associated with LDs) that could provide fatty acids to fuel DMV growth. These features are perfectly suited to refurbish the DMVs with lipids synthesized in the ER but to exclude 'undesired' ER proteins.
We found that NSP6 zippers the ER membrane through homodimerization and that NSP6(ΔSGF) (which underwent convergent evolution in the Alpha, Beta, Gamma, Eta, Iota and Lambda VOCs) has a higher ER-zippering activity. Notably, the recent and highly infectious BA.2 variant also contains the NSP6(ΔSGF) deletion18. The deletion is found in the second and longest luminal loop of NSP6, hosting a consensus O-glycosylation motif (LSGF: 105–108), which could act as a spacer that forms luminal bridges. Thus, the SGF deletion could convey higher zippering activity by shortening the ‘spacer’ and/or by preventing its O-glycosylation.
Our findings on NSP6 and its key role in replication organelle biogenesis provide a testable target that is easily amenable to screens for antiviral agents and that is applicable to a wide range of coronaviruses.
Methods
Reagents and antibodies
The following antibodies were used: mouse monoclonal anti-HA (BioLegend, 901503, dilution 1:600 for immunofluorescence (IF) and 1:1,500 for western blot (WB)), rabbit polyclonal anti-HA (Sigma-Aldrich, H6908, dilution 1:200 for IF), goat polyclonal anti-HA (Bethyl, A190-138A, dilution 1:600 for IF), rabbit polyclonal anti-actin (Sigma-Aldrich, A2066, dilution 1:10,000 for WB), rabbit polyclonal anti-NSP6 (ProSci, 9177, dilution 1:200 for IF and 1:1,000 for WB), sheep anti-NSP3 (The University of Dundee, DA126, dilution 1:100 for IF and 1:1,000 for WB), rabbit polyclonal ADRP/Perilipin 2 (Proteintech, 15294-1-AP, dilution 1:200), rabbit monoclonal anti-DFCP1 (Cell Signaling, 38419, dilution 1:1,000 for WB), mouse monoclonal anti-Flag (Sigma-Aldrich, F1804, dilution 1:400 for IF and 1:1,500 for WB), goat polyclonal anti-Flag (Bethyl, A190-101A, dilution 1:200 for IF), mouse monoclonal anti-c-Myc (Santa Cruz, sc-40, dilution 1:200 for IF), mouse monoclonal anti-GAPDH (Santa Cruz, sc-32233, dilution 1:1,000 for WB), mouse monoclonal anti-LAMP1 (Hybridoma Bank, H4A3, dilution 1:200 for IF), rabbit monoclonal anti-EEA1 (BD Biosciences, 610456, dilution 1:1,000 for IF), sheep anti-human anti-TGN46 (BioRad, AHP500GT, dilution 1:750 for IF), rabbit polyclonal anti-GFP (Abcam, ab6556, dilution 1:250 for IF), mouse monoclonal anti-GFP (Santa Cruz, sc-9996, dilution 1:2,000 for WB), mouse monoclonal anti-mCherry (Abcam, ab125096, dilution 1:2,000 for WB), mouse monoclonal anti-V5 (Thermo Fisher Scientific, R960-25, dilution 1:200 for IF and 1:1,000 for WB), rabbit polyclonal anti-LC3 (Novus Biologicals, NB100-2220, dilution 1:200 for IF), mouse monoclonal anti-dsRNA (Scicons, 10010500, dilution 1:10 for IF), DAPI (Sigma-Aldrich, D9542, dilution 1:10,000 for IF), rabbit 1.4-nm gold-conjugated Fab’ fragment (Nanoprobes, 2004, dilution 1:50), mouse 1.4-nm gold-conjugated Fab’ fragment (Nanoprobes, 2002, dilution 1:50), Alexa Fluor-546 FluoroNanogold anti-mouse Fab’ (7402, dilution 1:50) and Alexa Fluor-488-568-647 (Invitrogen, diluted 1:400), horseradish peroxidase (HRP)-conjugated goat anti-mouse or anti-rabbit IgG antibody (1:8,000, Merck Millipore, 401215 or 401315, respectively). Anti-GM130 (1:1,000 for IF) and anti-VAP-A (1:300 for IF) were produced in our laboratory as previously described31,32
BODIPY 493/503 (4,4-difluoro-1,3,5,7,8-pentamethyl-4-bora-3a,4a-diaza-s-indacene), β-BODIPY FL C12-HPC (2-(4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-dodecanoyl)-1-hexadecanoyl-sn-glycero-3 phosphocholine) and BODIPY 558/568-DA-C12 (4,4-difluoro-5-(2-thienyl)-4-bora-3a,4a-diaza-s-indacene-3-dodecanoic acid were purchased from Thermo Fisher Scientific (D3922, D3792 and D3835, respectively). Oil Red O solution was purchased from Merck (102419). K22 (N-[(1Z)-1-[[4-(4-bromophenyl)-4-hydroxy-1-piperidinyl]carbonyl]-2-phenylethenyl]-benzamide) was purchased from Cayman Chemical, the DGAT-1 inhibitor A922500 (A1737), wortmannin (3144), delipidated serum (S5394) and doxycycline hydrochloride (8D3447) from Sigma-Aldrich and the VPS34 specific inhibitor SAR405 from MedChemExpress (HY-12481). Puromycin dihydrochloride was purchased from Calbiochem (540411). For 35S-methionine/cysteine labelling, the EasyTag protein labelling mix (772007MC) was purchased from PerkinElmer. Unless otherwise stated, all other chemicals were purchased from Sigma-Aldrich.
Plasmid constructs
All NSP constructs were made with the Gateway system (Thermo Fisher Scientific) using a modified pCDNA3.1 vector (containing a HA, Flag, Myc, GFP or mCherry tag) for amino-terminal tagging, a modified pCDNA5/FRT/TO vector (containing 3×Flag) for carboxy-terminal tagging, unmodified pCDNA5/FRT/TO to clone untagged NSP6, and pLTD-Flag or pLTD-HA for stable doxycycline-inducible NSP6-expressing cell lines. All Gateway vectors were provided by P. Grumati. The donor plasmids were pDONR207 SARS-CoV-2 NSP3, pDONR223 SARS-CoV-2 NSP4 and pDONR223 SARS-CoV-2 NSP6 from Wuhan-Hu-1 SARS-CoV-2 (gifts from F. Roth, Addgene plasmids 141257, 141258 and 141260, respectively)33. For carboxy-terminal tagging of NSP6, the stop codon was removed using the oligo pairs NSP6 ns(+)/NSP6 ns(−) (Supplementary Table 2) with the Agilent QuikChange kit. The Agilent QuikChange kit and the oligos described in Supplementary Table 2 were used to make the following NSP6 N-terminally tagged mutant constructs: NSP6(1–157) (amino acids 1–157); NSP6-C80 (amino acids 211–290); the mutants in the amphiphilic alpha helix NSP6(F220Q/T222W) and NSP6-C80(F220Q/T222W); and the VOC mutant constructs NSP6(ΔSGF) and NSP6(ΔSGF)–NSP7.
The NSP6–NSP7 sequence was synthesized with flanking attB sequences by Thermo Fisher Scientific (Supplementary Table 1), a V5 tag was added to NSP7 by PCR, and the amplicon was cloned into the Gateway vector pDONR223 and recombined with destination vector pCDNA3.1 containing HA to produce pHA-NSP6-NSP7-V5.
The IBV (avian infectious bronchitis virus, strain M41) NSP6 sequence (corresponding to Uniprot P0C6Y3 from position 3089 to 3381), optimized for human expression and synthesized with flanking attB sequences by Thermo Fisher Scientific (Supplementary Table 1), was cloned into the Gateway vector pDONR223 and recombined in Flag-containing Gateway destination vectors at the amino or carboxy terminus. Oligos NSP6-IBV ns(+)/NSP6-IBV ns(−) (Supplementary Table 2) were used to remove the stop codon for the carboxy terminal-tagged construct.
mCherry-DFCP1 was a gift from D.-H. Kim (Addgene plasmid 86746). pEGFP-ATF6 was a gift from R. Prywes (Addgene plasmid 32955). mCherry-Calreticulin-N-16 (M. Davidson, Addgene plasmid 55006), pLenti-X1-Neo-GFP-ATL2 (J. Corn, Addgene plasmid 109020), pEGFPC-DFCP1 and pRUBY-N1-KDEL were provided by P. Grumati. pEGFP-RAB18 was a gift from M. Scidmore (Addgene plasmid 4955).
The Agilent QuikChange kit and the oligos described in Supplementary Table 2 were used to make the following mCherry–DFCP1 mutant constructs: DFCP1(Δ1–416) (lacking the amino terminus); DFCP1(W543A) (point mutation in the ER domain) and DFCP1(C654S/C770S) (mutations in the double FYVE domain; unable to bind PtdIns3P).
GST-tagged DFCP1 was constructed by amplifying the coding sequence from mCherry–DFCP1 with oligos DFCP1-p223(+)/DFCP1-p223(−) and cloning into the Gateway vector pDONR223 and subsequently into the Gateway vector pET60.
pEYFPC3-Cb5, constructed as described12 using YFP instead of mCherry, and pEGFP-VAP-A were made in our laboratory. pEGFP-ERGIC53 and p-KDELR-EGFP were gifts from A. Luini.
BP clonase and LR clonase for Gateway cloning were purchased from Thermo Fisher Scientific. All other reagents for molecular biology were purchased from New England Biolabs.
Cell culture, transfection and RNA interference
HeLa cells were obtained from ATCC and cultured as previously described12. Calu-3 cells (human lung adenocarcinoma), a gift from L. J. Galietta, were cultured in DMEM F-12 (Gibco), supplemented with 10% fetal bovine serum (Euroclone) 100 IU ml−1 penicillin and 100 µg ml−1 streptomycin (Thermo Fisher Scientific) and 2 mM l-glutamine (Thermo Fisher Scientific) in a humidified incubator at 37 °C and 5% CO2. Cell lines were routinely tested for mycoplasma (Biological Industries). Cells were transfected with plasmids using either TransIT-LT1 (Mirus Bio) for HeLa cells or Lipofectamine LTX and PLUS Reagent (Thermo Fisher Scientific) for Calu-3 according to the manufacturer’s instructions. Expression was maintained for 16–24 h before processing unless otherwise stated. For RNA interference, HeLa and Calu-3 cells were mock-treated or treated with DFCP1 siRNA (50 nM) for 96 h using Lipofectamine RNAiMAX (Thermo Fisher Scientific) for direct transfection. The siRNA sequences used in this study are listed in Supplementary Table 2.
Generation of HeLa Flag–NSP6 and HA–NSP6 doxycycline-inducible stable lines
To generate stably expressing clones, HeLa cells were transfected with the plasmids pLTD-Flag-NSP6, pLTD-Flag-NSP6(ΔSGF), pLTD-HA-NSP6 or pLTD-HA-NSP6(ΔSGF) and selected with complete medium containing 3 μg ml−1 puromycin (Calbiochem). Single-cell cultures were isolated from the mixed populations and protein expression was probed and induced with 1 μg ml−1 doxycycline (Sigma-Aldrich) at different time points, as indicated. Samples were then processed by immunofluorescence analysis. All the cell lines generated in this study were authenticated through western blot and immunofluorescence.
SARS-CoV-2 infection and assays
SARS-CoV-2 infection, virus titration and cell death assay through the activity of lactate dehydrogenase (LDH) were performed as elsewhere described24. For immunofluorescence experiments, Calu-3 cells were seeded on coverslips, left untreated or pre-treated for 2 h with K22 or with the DGAT-1 inhibitor A922500 at different concentrations, as indicated in the figures. Cell number and cell viability after treatment with either K22 or A922500 were assessed by crystal violet staining, cell morphology analysis or LDH assay. No cytostatic or cytotoxic effect of the drugs was observed at the concentrations used. For immunofluorescence experiments and drug treatments, Calu-3 cells were seeded on coverslips and infected with SARS-CoV-2 early lineage (SARS-CoV-2/human/BRA/RJ01/2020, GenBank accession no. MT710714) at a multiplicity of infection (MOI) of 0.01 for 48 h. Infected cells were fixed with 3.7% formaldehyde and processed for immunofluorescence as described24. For comparative analyses of NSP3–NSP6 proximity, cells were similarly infected with early lineage and Gamma variant (hCoV-19/Brazil/AM-L70-71-CD1739/2020, GISAID ID: EPI_ISL_1060902) SARS-CoV-2 at a MOI of 0.01 for 48 h.
For EM experiments, Calu-3 cells were infected with early lineage B.1 (hCoV-19/Italy/CAM-INMI-32803-66/2020, GISAID ID: EPI_ISL_493333) or Gamma variant (hCoV-19/Italy/CAM-IZSM-RD020483D54/2021, GISAID ID: EPI_ISL_2933105) SARS-CoV-2 strains at an MOI of 10 for 24 h. SARS-CoV-2-infected Calu-3 cells were processed for EM as described below. All procedures related to virus culture were handled at a biosafety level 3 (BSL3) multi-user facility, according to World Health Organization (WHO) guidelines.
Drug treatments
Flag–NSP6- and mCherry–DFCP1-transfected cells were treated with either 100 nM wortmannin or 1 µM VPS34 inhibitor SAR405 for 3 h, then processed for immunofluorescence. For K22 treatment, cells were transfected and after 30 min dimethyl sulfoxide (DMSO) or 40 µM K22 were added.
Recombinant proteins and pull-down and co-immunoprecipitation experiments
All recombinant proteins were purified from Escherichia coli Rosetta DE3 cells (Merck). GST-tagged DFCP1 from plasmid pET60 and GST alone from plasmid GEX-4T2 (GE Healthcare) were expressed as described34. For pull-down experiments, 3 mg of cellular lysates from HA-NSP6-transfected HeLa cells was incubated with GST–DFCP1 or GST alone (0.1 μM) overnight at 4 °C in 950 μl binding buffer (25 mM Tris pH 7.4, 150 mM NaCl, 0.1% Triton-X-100, 0.1% NP-40, 1 mM EDTA and protease inhibitors). Glutathione beads were added, incubated for 1 h at 4 °C, washed four times with incubation buffer and twice with a similar buffer without detergents, eluted and analysed by SDS–PAGE.
For co-immunoprecipitation experiments, 1.7 mg of cellular lysate from cells mock-transfected or co-transfected with HA–NSP6 together with GFP–NSP6, Flag–NSP6, GFP–ERGIC53, GFP–atlastin-2 or GFP–NSP6(1–157), or co-transfected with HA–NSP6(ΔSGF) and GFP–NSP6(ΔSGF), were incubated with appropriate antibody-conjugated beads (HA, Flag and GFP). After overnight incubation at 4 °C in 750 μl binding buffer, samples were washed five times with binding buffer and once with a similar buffer without detergents, eluted and analysed by SDS–PAGE. To evaluate co-immunoprecipitation efficiency, a total of three independent experiments were analysed. The co-immunoprecipitated GFP–NSP6 signal was divided by the GFP–NSP6 signal in the input and normalized by the signal of the immunoprecipitated primary antigen (HA). Co-immunoprecipitation efficiency was reported as mean ± s.e.m. of co-immunoprecipitated GFP–NSP6(ΔSGF) compared to GFP–NSP6.
Detergent extraction
HeLa cells transfected with Flag–NSP6, NSP6–Flag or Flag–NSP6(ΔSGF) were lysed in buffer (25 mM Tris pH 7.4, 150 mM NaCl, 1 mM EDTA with protease and phosphatase inhibitor cocktails) containing increasing concentrations of Triton-X-100 and NP-40 (1:1) and centrifuged at 13,200 rpm for 10 min. The pellet was resuspended in the same volume as the supernatant and equal volumes were subjected to western blot analysis using an anti-Flag antibody.
Metabolic radiolabelling
For metabolic labelling, wild-type HeLa cells or the pLTD-HA-NSP6 or pLTD-HA-NSP6-ΔSGF stable cell lines were induced with doxycycline (1 μg ml−1) for 13 h, incubated for 30 min with methionine/cysteine-free medium (21013024, Gibco), and then incubated for 1 h at 37 °C with 50 μCi ml−1 35S-methionine/cysteine (PerkinElmer) in the same medium. The cells were then washed three times with complete medium and further incubated for different times at 37 °C in complete medium. Doxycycline (1 μg ml−1) was included in all media. After cell lysis, proteins were immunoprecipitated with anti-HA affinity beads, and analysed by SDS–PAGE gel autoradiography (using a Typhoon Imager, Image QuantTool, GE Healthcare) of the immunoprecipitates to measure protein stability followed by immunoblot using anti-HA to measure total protein levels.
Western blot analysis
Western blot analysis and densitometry were performed as previously described34. Samples containing NSP6 were mixed with sample buffer (100 mM Tris pH 6.8, 25% glycerol, 2% SDS, 0.01% bromophenol blue and 10% 2-mercaptoethanol), but were not boiled before loading.
Immunofluorescence analysis
Immunofluorescence analysis was performed as previously described12.
Digitonin and Triton-X-100 permeabilization
HeLa cells transfected with Flag–NSP6 or NSP6–Flag were grown on coverslips and fixed with 4% PFA for 10 min, washed three times with buffer A (20 mM PIPES pH 6.8, 137 mM NaCl and 2.7 mM KCl) and permeabilized with 20 µM digitonin (Calbiochem) diluted in buffer A for 5 min. Coverslips were blocked for 30 min with blocking solution (5% FBS (v/v) and 50 mM NH4Cl in buffer A) without any additional permeabilizing agent and incubated with primary anti-Flag and anti-TGN46 antibodies diluted in blocking solution. The TGN46 antibody was raised against a luminal portion of the protein that is thus not accessible after digitonin permeabilization. This represents a control in that only the plasma membrane has been permeabilized. Coverslips were washed with buffer A and incubated with fluorochrome-conjugated secondary antibodies (Alexa Fluor-488 for Flag and Alexa Fluor 568 for TGN46 in buffer A) for 1 h at room temperature. After incubation, cells were fixed with 2% PFA for 5 min and washed once with 50 mM NH4Cl in PBS. Coverslips were subsequently permeabilized with 0.1% Triton-X-100 in PBS for 5 min. Cells were then blocked with blocking solution (0.05% saponin, 0.5% BSA and 50 mM NH4Cl in PBS) and incubated with the same primary antibodies used in the first step. Coverslips were then washed with PBS and incubated with fluorochrome-conjugated secondary antibodies (Alexa Fluor 405 for Flag and Alexa Fluor 633 for TGN46 in PBS) for 1 h at room temperature. The TGN46 epitope becomes accessible to the primary antibody under these conditions, confirming selective permeability and identifying luminal epitopes.
LD staining and assays
LDs were stained by adding 0.5 μM BODIPY 493/503 (Thermo Fisher Scientific) to the fluorochrome-conjugated secondary antibody mix for 30 min after fixation and processed as for immunofluorescence analysis.
To monitor lipid transfer from LDs to DMVs we followed the protocol described previously30. In brief, BODIPY 558/568-DA-C12 at a final concentration of 1 μM was added for 16 h to the culture medium of HeLa cells transfected with GFP–NSP4/HA–NSP3 or GFP–NSP4/HA–NSP3/Flag–NSP6. Cells were then washed and incubated with DMEM supplemented with delipidated serum (1%) for an additional 6 h. Coverslips were fixed and processed as described above. NSP4 puncta were identified by using the ‘Analyze particles’ tool of Fiji (ImageJ) software, and the fluorescence mean intensity of Bodipy-DA-C12 for each particle was determined. Particles with values equal or higher than a similar area of the ER were defined as ‘positive’ particles. The percentage of NSP4 Bodipy-DA-C12-positive particles was calculated for each cell.
Confocal microscopy and image analyses
Cells were imaged using a Plan-Apochromat 100×/1.4 oil objective on a Zeiss LSM800 or LSM880 confocal system equipped with an AiryScan module and controlled by the Zen blue software. Fluorescence images presented are representative of images collected from at least three independent experiments, unless otherwise stated (see 'Statistics and Reproducibility' for further details). The images used for phenotype quantification were acquired with the same parameters (that is, digital gain, laser power and magnification) and processed with Fiji (ImageJ; National Institues of Health (NIH)) software. Brightness and contrast were adjusted with Adobe Photoshop, and figure panels were assembled with Adobe Illustrator.
Quantification of the number and area of structures
NSP6, NSP4, LC3 and LD structures were analysed using the ‘Analyze particle’ function to determine their number per cell. For each experiment, images were acquired below saturation limit and the same threshold was chosen and applied to all of them. For the calculation of the size of the structures the 'Analyze particle' function was used, setting ‘Area’ as measurement.
Distribution of NSP4 puncta
To calculate the distribution of NSP4 puncta in each cell, the ‘Analyze particle’ function was used, considering a particle size between 0.1 and infinity and choosing the centre of mass as a reference for measurement. X and Y coordinates for each NSP4 puncta were obtained and plotted. A four-quadrant subdivision was applied to the images using the XY coordinates of the centre of mass as the axis origin. The relative abundance of the NSP4 puncta for each quadrant is expressed as a percentage of the total identified structures for each cell.
Relative distribution of the NSP6 protein
To measure the cellular distribution of NSP6 fluorescence, the integrated density of NSP6 in NSP6 structures was calculated over the integrated density of total NSP6 in the whole cell.
Cells with comparable levels of total integrated fluorescence intensity were analysed for each time point. Results were expressed as a percentage of the fluorescent NSP6 signal present in the NSP6 structures over the total fluorescence.
Recruitment to NSP6 structures
The fraction of VAP-A or NSP6(1–157) associated with NSP6-positive structures was measured as the ratio between the integrated density of each protein on the NSP6 structures and the integrated density in the whole cell.
Co-localization between NSP6 and DFCP1
Co-localization between NSP6 and wild-type or mutant DFCP1 was calculated using the JACoP plug-in35.
Distance between particles
The relative distance between objects was determined with the DiAna plug-in36. In brief, channels were thresholded and then segmented. For LD distance from NSP4 and NSP6 in transfected cells shown in Fig. 4b, edge–edge distances between particles were measured in the whole cell. No values were excluded. In addition, for selected images including the one in Fig. 4a we applied the Shuffle function36 as shown in Extended Data Fig. 9f. In brief, this function redistributes the objects in a channel in a random manner; then the distances between objects of the randomized channel to the closest object in the second channel from the original image are measured. The distribution of these distances is represented as the mean (red line) flanked by 95% confidence intervals (green lines). The distribution of the distances measured between the objects in the two channels from the original images is plotted (blue line). If this distribution falls outside the confidence interval of the distance obtained for shuffled images, the distance is considered as statistically significant (P < 0.05).
For the proximity between LDs and dsRNA or NSP6 in Extended Data Fig. 9d, and the proximity between dsRNA and NSP6 in Extended Data Fig. 7i, in infected cells, the edge–edge distance was analysed and structures closer than 250 nm (in all directions) were considered as associated structures. To calculate the distance between NSP3- and NSP6-positive structures in infected cells in Fig. 3b both centre–centre and edge–edge distances were measured.
Measurements of NSP6 fluorescence intensity
HeLa cells expressing Flag-tagged NSP6 were fixed and processed for immunofluorescence. Cells with similar expression were acquired using the same parameters and processed with the Fiji (ImageJ) software. The integrated density of each cell was measured.
Electron microscopy
For pre-embedding IEM, the cells were fixed, permeabilized and labelled as described previously37. In brief, the cells were fixed with a mixture of 4% paraformaldehyde (PFA) and 0.05% glutaraldehyde prepared in 0.2 M HEPES buffer for 10 min (room temperature) and then with 4% PFA alone for 30 min (room temperature), followed by incubation with blocking/permeabilizing solution (0.5% bovine serum albumin (BSA), 0.1% saponin and 50 mM NH4Cl in PBS) for 30 min.
Cells were incubated with a primary anti-HA monoclonal antibody (1:600, BioLegend) diluted in blocking/permeabilizing solution overnight and then a secondary anti-mouse antibody (1.4-nm gold-conjugated Fab’ fragment diluted 1:50, Nanoprobes) was added for 2 h. The GoldEnhance EM kit (from Nanoprobes) was used to enhance ultrasmall gold particles. For double labelling of cells expressing HA–NSP3, mCherry–NSP4 and GFP–NSP6, enhancement with the anti-HA antibody was performed for 3 min and then a primary anti-GFP polyclonal rabbit antibody (1:250, Abcam) was added and processed as above using a secondary anti-rabbit antibody (1.4-nm gold-conjugated Fab’ fragment diluted 1:50, Nanoprobes) for 2 h, followed by gold enhancement for an additional 3 min. The longer enhancement time for the anti-HA detection causes the formation of larger gold particles (clusters) with an irregular shape that distinguishes HA–NSP3 from the smaller GFP–NSP6 signals in doubly transfected cells.
For conventional EM, the cells were fixed with 1% GA prepared in 0.2 M HEPES buffer for 30 min (RT).
Cells prepared for IEM or conventional EM were scraped, pelleted, post-fixed in OsO4 and uranyl acetate, dehydrated, embedded in Epon and polymerized at 60 °C for 72 h. For each sample, thin sections were cut using a Leica EM UC7 ultramicrotome (Leica Microsystems). EM images were acquired from thin sections using a FEI Tecnai-12 electron microscope (FEI) equipped with a VELETTA CCD digital camera (Soft Imaging Systems). Morphometric analysis of the structures of interest was performed using iTEM software (Olympus).
CLEM
HeLa cells were transfected with HA–NSP6 or HA–NSP6(ΔSGF), or they were co-transfected with HA–NSP3/mCherry–NSP4/GFP–NSP6 or HA–NSP3/mCherry–NSP4/Myc–NSP6 where indicated. Transfected cells were treated or not with 40 µM K22 30 min after transfection. After overnight expression, cells were fixed as for IEM and then labelled with an anti-HA antibody followed by detection with a secondary Alexa Fluor-546 FluoroNanogold anti-mouse Fab’. The structures of interest carrying different proteins were visualized by confocal microscopy using a Zeiss LSM800 station and fluorescent images were recorded. Then the cells were post-fixed, dehydrated, embedded in Epon and polymerized as described above. Serial 60-nm sections were cut and analysed using a FEI Tecnai-12 electron microscope. The same cell and structures of interest obtained by confocal microscopy were identified on EM images using Zen Connect software (Zeiss).
Electron tomography
Epon sections (250 nm thick) were collected on Formvar carbon-coated slot grids and analysed using a Tecnai G2 Spirit BioTwin electron microscope (FEI) equipped with an automated tomography stage. The single tilt series of images were acquired in a range of −65° to +65° (at 1° intervals) using Xplore 3D TEM Tomography software (FEI) at 40,000× magnification unless otherwise stated. Tilt series were used with the open-source IMOD software to generate tomograms. At least 10 tomograms were analysed per experimental condition. For 3D reconstruction, the surfaces of DMVs and surrounding ER membranes were rendered using the IMOD software.
FLIM measurements, FRAP and FLIP analysis
FLIM–FRET analysis of GFP–NSP6 alone and in combination with mCherry–NSP6, mCherry or mCherry–DFCP1, and of mCherry–NSP6 with GFP–NSP6(1–157), GFP–ERGIC and GFP–atlastin-2, was performed as previously described12. FLIM data analysis was performed using SymPhoTime 64 (Picoquant). For live-cell imaging of the NSP6 structures, cells were plated in glass-bottomed dishes (MatTek), transfected with the fluorescently tagged protein constructs or incubated with β-BODIPY FL C12-HPC (1 μM) for 16 h, and imaged with an LSM800 microscope (Zeiss) fitted with 488- and 561-nm argon laser lines, using a 63× Plan-Apochromat NA 1.4 DIC oil immersion objective. During imaging, cells were maintained in complete culture medium in a humidified atmosphere at 37 °C. Fluorescence images presented are representative of cells imaged in at least three independent experiments and were processed with Fiji (ImageJ; NIH) software.
FRAP experiments and time-lapse laser-scanning confocal microscopy were performed as described12. In brief, a single NSP6 structure was acquired five frames before bleaching (6 s per frame). Bleaching was performed with 100% power of the 488 laser for 10 iterations. Recovery was monitored for 600 s after the bleaching event. At least 30 independent structures were analysed for each condition in three different experiments. Data were exported using Zen software (Zeiss) and corrected for bleaching by dividing the fluorescence intensity of the bleached area by that of an unbleached area. Bleaching was minimal during the time course of recovery (between 0 and 10%): where bleaching exceeded 10%, the recovery sequences were discarded. Quantification of GFP–NSP6 and GFP–NSP6(ΔSGF) dissociation from membranes was measured in living cells by FLIP. FLIP was performed in cells expressing each GFP-tagged protein by bleaching iteratively (100 times, with intervals of 6 s between frames) the GFP-associated fluorescence in the entire cell area except for a region of interest (ROI) containing NSP6 structures. The ROI usually accounted for 10–15% of the total cell area. The relative fluorescence intensity of single structures expressed as a percentage of pre-bleaching fluorescence was plotted as mean values ± s.d. A slowdown of the FLIP-induced decay curves of GFP–NSP6(ΔSGF) from the structures was observed indicating an increase in GFP–NSP6(ΔSGF) association with membranes.
EM quantification
The percentage of normal and zippered ER (or NE) surface was quantified in random thin sections from pellets of NSP6-transfected HeLa cells using morphometric grids with the iTEM software (Olympus SIS). Quantification of gold particles in thin sections from HeLa cells expressing HA–NSP6 or HA–NSP6(ΔSGF) and immuno-gold labelled for HA was performed with the touch count tool of the iTEM software. This quantification was further used as a measure of HA–NSP6 or HA–NSP6(ΔSGF) expression in each analysed cell to normalize the surface area of zippered ER for the expression level of the corresponding HA-tagged NSP6 protein. To assess the effect of NSP6 or NSP6(ΔSGF) on the organization of DMVs, tomograms of DMV clusters were used to quantify the following parameters: DMV diameter, shape factor (ratio between long and short axes), density (number per DMV cluster area), length of ER–DMV connections, number of DMVs per connection and overall number of ER–DMV connections per DMV cluster. DMV cluster was defined as a group of DMVs whose distance from the nearest neighbour does not exceed two average DMV diameters. All measurements in tomograms were done with the 3D Manager plug-in of the open-source Fiji software. The same tools were used to quantify the length of zippered DMV connectors in tomograms from Calu-3 cells infected with the early lineage B.1 or Gamma variant of SARS-CoV-2.
NSP6 protein topology
NSP6 topology modelling was performed using the Constrained Consensus TOPology prediction server (CCTOP, Institute of Enzymology). The amphipathic features of the α-helix were determined using HELIQUEST (http://heliquest.ipmc.cnrs.fr)16 and the mutations were introduced following the Genetic Algorithm-based module. The images and cartoons shown in Fig. 2a and Extended Data Fig. 11 were created with BioRender.com.
Phylogenetic analysis
The phylogenetic analysis of SARS-CoV-2 genomes deposited in the GISAID database (https://www.gisaid.org/) was performed on a set of 3,508 representative genomes sampled from December 2019 to July 2021, provided by Nextstrain18 (https://nextstrain.org/ncov/global). The percentages of genomes carrying the SGF deletion in the NSP6 protein were evaluated on samples deposited at GISAID up to 16 July 2021.
Statistics and reproducibility
Statistical analyses were performed using GraphPad Prism7 (GraphPad Software) or the R software environment for statistical computing (rstatix R package).
To test the normal distribution of the data and the homogeneity of variance across groups, the Shapiro–Wilk test and Levene’s test were used on the ANOVA residuals. When measured variables were normally distributed, the statistical significance of difference in measured variables between control and treated groups was determined by t-test or ANOVA followed by appropriate multiple comparison post-hoc tests depending on the experiment. When the measured variables were not normally distributed, non-parametric Mann–Whitney or Kruskal–Wallis tests were performed followed by appropriate multiple comparison post-hoc tests depending on the experiment.
All the experiments for which statistics was derived were performed three times with similar results; N indicates the number of experiments and n the number of total measurements or observations. All of the replicates performed were biological and not technical. Detailed information for each experiment is provided below.
The experiments shown in Extended Data Figs. 1a,e, 3h, 6a and 9c were repeated twice.
The experiments shown in Figs. 1b,c,g–i, 2i,j, 3d,e,f,i,n and 4c and Extended Data Figs. 1f–j, 2a–c, 3a,d, 4c–i, 5k,l, 6d,e,i,j,l,m, 7c–e, 8a,b,f–h, 9a,b,f–h and 10a,b were repeated three times.
The experiments shown in Extended Data Figs. 1c,d, 2d,e, 5c,m and 6c,f were repeated four times. The experiments shown in Figs. 1a, 3a and 4d and Extended Data Figs. 6g, 9i and 10c were repeated five times. The experiment shown in Extended Data Fig. 5b was repeated six times.
The experiments shown in Figs. 1e,f and 3c and Extended Data Figs. 5j and 6b were repeated ten times.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
Full scans for all western blots and autoradiographs are provided in Supplementary Fig. 1. The nucleotide sequences of synthetic IBV NSP6 and NSP6–NSP7 used in this study are provided in Supplementary Table 1. The oligonucleotides, siRNAs and primers used in this study are provided in Supplementary Table 2. Raw data supporting the findings of this study are deposited in Zenodo and are publicly available at https://doi.org/10.5281/zenodo.5929088. Raw EM data, including tilt series and reconstructed 3D tomograms, were deposited in the Electron Microscopy Data Bank (EMDB) and the Electron Microscopy Public Image Archive (EMPIAR) public databases with accession codes EMD-14179 and EMPIAR-10935, respectively. Source data are provided with this paper.
References
Hartenian, E. et al. The molecular virology of coronaviruses. J. Biol. Chem. 295, 12910–12934 (2020).
Cortese, M. et al. Integrative imaging reveals SARS-CoV-2-induced reshaping of subcellular morphologies. Cell Host Microbe 28, 853–866.e5 (2020).
Angelini, M. M., Akhlaghpour, M., Neuman, B. W. & Buchmeier, M. J. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio 4, e00524–13 (2013).
Oudshoorn, D. et al. Expression and cleavage of Middle East respiratory syndrome coronavirus nsp3-4 polyprotein induce the formation of double-membrane vesicles that mimic those associated with coronaviral RNA replication. mBio 8, e01658-17 (2017).
Wolff, G., Melia, C. E., Snijder, E. J. & Bárcena, M. Double-membrane vesicles as platforms for viral replication. Trends Microbiol. 28, 1022–1033 (2020).
Snijder, E. J. et al. A unifying structural and functional model of the coronavirus replication organelle: tracking down RNA synthesis. PLoS Biol. 18, e3000715 (2020).
Klein, S. et al. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 11, 5885 (2020).
Ogando, N. S. et al. SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J. Gen. Virol. 101, 925–940 (2020).
Peacock, T. P., Penrice-Randal, R., Hiscox, J. A. & Barclay, W. S. SARS-CoV-2 one year on: evidence for ongoing viral adaptation. J. Gen. Virol. 102, 001584 (2021).
Romero, P. D. et al. The emergence of SARS-CoV-2 variant lambda (C.37) in South America. Microbiol. Spectr. 9, e0078921 (2021).
Cottam, E. M. et al. Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy 7, 1335–1347 (2011).
Venditti, R. et al. Molecular determinants of ER–Golgi contacts identified through a new FRET-FLIM system. J. Cell Biol. 218, 1055–1065 (2019).
Dobson, L., Reményi, I. & Tusnády, G. E. CCTOP: a Consensus Constrained TOPology prediction web server. Nucleic Acids Res. 43, W408–W412 (2015).
Baliji, S., Cammer, S. A., Sobral, B. & Baker, S. C. Detection of nonstructural protein 6 in murine coronavirus-infected cells and analysis of the transmembrane topology by using bioinformatics and molecular approaches. J. Virol. 83, 6957–6962 (2009).
Oostra, M. et al. Topology and membrane anchoring of the coronavirus replication complex: not all hydrophobic domains of nsp3 and nsp6 are membrane spanning. J. Virol. 82, 12392–12405 (2008).
Gautier, R., Douguet, D., Antonny, B. & Drin, G. HELIQUEST: a web server to screen sequences with specific α-helical properties. Bioinformatics 24, 2101–2102 (2008).
Lundin, A. et al. Targeting membrane-bound viral RNA synthesis reveals potent inhibition of diverse coronaviruses including the Middle East respiratory syndrome virus. PLoS Pathog. 10, e1004166 (2014).
Hadfield, J. et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 34, 4121–4123 (2018).
Twu, W. I. et al. Contribution of autophagy machinery factors to HCV and SARS-CoV-2 replication organelle formation. Cell Rep. 37, 110049 (2021).
Tabata, K. et al. Convergent use of phosphatidic acid for hepatitis C virus and SARS-CoV-2 replication organelle formation. Nat. Commun. 12, 7276 (2021).
Holwerda, M., V’kovski, P., Wider, M., Thiel, V. & Dijkman, R. Identification of an antiviral compound from the pandemic response box that efficiently inhibits SARS-CoV-2 infection in vitro. Microorganisms 8, 1872 (2020).
Lee, J. Y. et al. Absolute quantitation of individual SARS-CoV-2 RNA molecules provides a new paradigm for infection dynamics and variant differences. eLife 11, e74153 (2022).
Thorne, L. G. et al. Evolution of enhanced innate immune evasion by SARS-CoV-2. Nature 602, 487–495 (2022).
Dias, S. S. G. et al. Lipid droplets fuel SARS-CoV-2 replication and production of inflammatory mediators. PLoS Pathog. 16, e1009127 (2020).
Li, D. et al. The ER-localized protein DFCP1 modulates ER–Lipid droplet contact formation. Cell Rep. 27, 343–358 (2019).
Gao, G., Sheng, Y., Yang, H., Chua, B. T. & Xu, L. DFCP1 associates with lipid droplets. Cell Biol. Int. 43, 1492–1504 (2019).
Herker, E., Vieyres, G., Beller, M., Krahmer, N. & Bohnert, M. Lipid droplet contact sites in health and disease. Trends Cell Biol. 31, 345–358 (2021).
Ridley, S. et al. FENS-1 and DFCP1 are FYVE domain-containing proteins with distinct functions in the endosomal and Golgi compartments. J. Cell Sci. 114, 3991–4000 (2001).
Axe, E. L. et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182, 685–701 (2008).
Rambold, A. S., Cohen, S. & Lippincott-Schwartz, J. Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev. Cell 32, 678–692 (2015).
Marra, P. et al. The GM130 and GRASP65 Golgi proteins cycle through and define a subdomain of the intermediate compartment. Nat. Cell Biol. 3, 1101–1113 (2001).
Jansen, M. et al. Role of ORPs in sterol transport from plasma membrane to ER and lipid droplets in mammalian cells. Traffic 12, 218–231 (2011).
Kim, D. K. et al. A comprehensive, flexible collection of SARS-CoV-2 coding regions. G3 10, 3399–3402 (2020).
Venditti, R. et al. The activity of Sac1 across ER–TGN contact sites requires the four-phosphate-adaptor-protein-1. J. Cell Biol. 218, 783–797 (2019).
Bolte, S. & Cordelières, F. P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232 (2006).
Gilles, J. F., Dos Santos, M., Boudier, T., Bolte, S. & Heck, N. DiAna, an ImageJ tool for object-based 3D co-localization and distance analysis. Methods 115, 55–64 (2017).
Polishchuk, E. V. & Polishchuk, R. S. Pre-embedding labeling for subcellular detection of molecules with electron microscopy. Tissue Cell 57, 103–110 (2019).
Acknowledgements
We thank P. P. M. Rede, C. B. W. G. Gripp, E. Van Anken, C. Machamer,R. De Francesco, M. Chiara, A. Ballabio, G. Diez Roux, A. Luini, R. Sitia, C. Settembre, D. Cacchiarelli, A. Grimaldi and P. Grumati for discussions and A. Iuliano for assistance with the statistical analysis. M.A.D.M. acknowledges the support of Telethon (grant TGM16CBDM13), the Italian Association for Cancer Research (grant IG2013_14761), the European Research Council Advanced Investigator grant 670881 (SYSMET), the University of Naples Federico II (grant STAR Plus 2020 Linea 1) and the Italian Ministry of University and Research (PRIN, 2020PKLEPN). R.S.P. acknowledges the support of Telethon (grant TGM16CBDM09). R.V. acknowledges the University of Naples Federico II (grant STAR 2017 Linea 1) and the Italian Association for Cancer Research (grant MFAG 2020, code 25174). P.T.B. and T.M.L.S. acknowledge the support of the Inova program Fiocruz, Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
Author information
Authors and Affiliations
Contributions
M.A.D.M. conceived the work. R.V. coordinated the experimental plan. R.V., S.R., A.M.G. and L.G. planned and analysed most of the experiments. E.V.P. and R.S.P. performed the EM, CLEM and tomography analyses. M.S. and G.D.T. developed plasmid constructs and provided technical support. A.M.G. and G.D.T. performed the protein studies. C.W. provided background data analysis. F.P. performed the VOC evolution analysis. V.C.S., S.S.G.D., J.C.S., T.M.L.S., P.T.B., G.F., M.V. and S.B. performed the SARS-CoV-2 studies. M.A.D.M. conceptualized the work and strategy and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 NSP6 requires a free C terminus to exert its membrane-deforming activity.
a, Fluoromicrographs of Calu-3 cells expressing YFP-Cb5 alone (leftmost panels) or in combination with C-terminally or N-terminally Flag-tagged NSP6, or untagged NSP6 (NSP6), as indicated, and immunostained with anti-Flag antibody (red). b, Expression analysis of NSP6-Flag (C-term) or Flag-NSP6 (N-term) in transfected HeLa cells. Left, representative fluorescence micrographs (anti-Flag antibody). Right, fluorescence intensity measurements. Single values are plotted, Means ± SEM are indicated. N = 3, n = 69 cells. ns, not significant. Two-tailed unpaired t-test with Welch’s correction. Lower panel, western blot of total protein lysates using an anti-Flag antibody; actin was used as a loading control. NT, non-transfected cells. c, Western blot of HeLa cells expressing HA-NSP6 or untagged NSP6, detected using anti-HA or anti-NSP6 antibody, as indicated. NT, non-transfected cells. d, HeLa cells transfected with Flag-NSP6 immunostained with anti-Flag, anti-LAMP1, anti-EEA1 or anti-LC3 antibodies. e, Fluoromicrographs of HeLa cells expressing YFP-Cb5 and either IBV NSP6-Flag (upper panels) or IBV-Flag-NSP6 (lower panels). Cells immunostained with anti-Flag antibody (red). f, IEM (anti-HA immuno-gold-labelling) of a HeLa cell expressing HA-NSP6. g, Magnification of the boxed area. Arrows show the regular single membrane of the ER cisterna; arrowheads indicate zippered membranes in the circular NSP6-positive structure. h, Single slices from a tomogram of a HeLa cell expressing HA-NSP6. Connections of circular zippered structures with the ER are shown by black arrows, and the connection of linear zippered membranes with the nuclear envelope is indicated by a white arrow (see Supplementary Video 1). i, Routine EM of a HeLa cell expressing HA-NSP6 and j, magnification of boxed area. The arrow indicates apposition of limiting membranes of an ER cisterna (asterisk) that then continues into the zippered ER domain (arrowheads). Western blots in b,c are representative of three independent experiments each. Scale bars, a, b, d, e, 10 µm; f, g, 100 nm; h, 230 nm; i, 200 nm; j, 100 nm.
Extended Data Fig. 2 The NSP6 compartment is accessible to ER membrane proteins with small luminal domains but not to membrane proteins with large luminal domains.
a, Fluoromicrographs of HeLa cells expressing CLRT-mCherry alone (left panel) or with GFP-NSP6 (right panel). b, Fluoromicrographs of HeLa cells expressing GFP-ATF6 or GFP-KDEL or GFP-ERGIC53 alone (left panel) or with mCherry-NSP6 (middle and right panels). c, Fluoromicrographs of HeLa cells expressing GFP-ATL2 alone (left panel), or with mCherry-NSP6 (middle panel), or expressing GFP-VAP-A (right panel). d, HeLa cells expressing GFP-KDELR alone (left panel) or with mCherry-NSP6 (middle and right panels). Small panels in (b–d), enlargements of boxed areas, arrowheads indicate co-localization in (d). e, Representative images of Hela cells expressing YFP-Cb5 alone or with mCherry-NSP6, as indicated. For FRAP analysis, individual NSP6 compartments (boxed) were photobleached and the fluorescence recovery was monitored. The small panels are representative frames (from a total of 100) showing time in seconds after the bleach (see Supplementary Video 3). See Fig. 1k for FRAP measurements. f, Representative images from FRAP experiments of Hela cells incubated with BODIPY C12-HPC (PC), without or with mCherry-NSP6 transfection, as indicated. Individual NSP6 compartments (boxed) were photobleached and the fluorescence recovery was monitored. The small panels are representative frames (from a total of 100) showing time in seconds after the bleach (see Supplementary Videos 2 and 3). Scale bars, 10 μm.
Extended Data Fig. 3 NSP6 undergoes homodimerization through the 1–157 region.
a, HeLa cells expressing C-terminal or N-terminal Flag-tagged NSP6 immunostained with anti-Flag antibody and an antibody against a luminal epitope of TGN46 after permeabilization with digitonin and subsequently with Triton-X-100 (see Methods). b, Model of the amphipathic helix of NSP6 (left panel) according to HELIQUEST (see Methods). Apolar residues are in yellow, polar residues and glycine have been given different colours. The arrow indicates the hydrophobic moment (µH = 0.409). Numbers indicate amino acid positions of the NSP6 protein. Right panel, model of the F220Q/T222W NSP6 mutant helix (µH = 0.191). Mutations that abolish the amphipathic character of the helix are in red. c, HeLa cells untransfected (left panel) or expressing Myc-NSP6 were immunostained for VAP-A or for Myc. Insets show the Myc-NSP6 signal. The number indicates the fraction of VAP-A associated with the NSP6 structures. Mean ± SD, N = 3, n = 74. d, HeLa cells expressing GFP-NSP6 F220Q/T222W mutant. e, f, g, Cell lysates (input) and immunoprecipitates (IP, with anti-HA or anti-Flag antibodies) from HeLa cells, untransfected or expressing the indicated NSPs were analysed by western blot with anti-HA, anti-Flag or anti-GFP antibodies as appropriate. Images are representative of three independent experiments. h, Fluoromicrographs of HeLa cells expressing GFP-NSP6(1–157) alone or with mCherry-NSP6. Scale bar, a, c, d, h, 10 µm.
Extended Data Fig. 4 K22 interferes with the formation of the NSP6 compartment.
a, Stably transfected Flag-NSP6 clone induced with doxycycline and treated with DMSO or K22 for 24 h. K22 reduced the number of NSP6 structures and resulted in elongated structures in a percentage of the cells (right panel, number of cells exhibiting these structures. Mean ± SD). The number of NSP6 structures in DMSO and K22-treated cells is plotted as single values. Mean ± SEM, N = 3, n = 90. Two-tailed unpaired t-test with Welch’s correction. b, FRAP analysis of mCherry-NSP6-structures (boxed) in cells treated with DMSO or K22. The small panels show time in seconds after the bleach. Graph, FRAP curves expressed as a % of time 0. Mean ± SD, N = 3, n = 45 structures. c, Cells expressing GFP-VAP-A alone (as a control) or with Flag-NSP6, treated with DMSO or with K22 for 16 h. d–f, Immuno-CLEM of HA-NSP6-expressing cells treated with K22 for 24 h. d, Fluoromicrograph showing NSP6 (anti-HA immunostaining with Alexa Fluor®546-FluoroNanogold secondary antibody) in elongated structures (arrows 1, 2) close to the nucleus (asterisk). e, EM section of the same cells (asterisks) shown in (d). Arrows 1 and 2 indicate overlap of the fluorescent and immuno-gold signals in the elongated zippered domains of the nuclear envelope (NE). f, Serial sections of the structure indicated by arrow 1 in panel (e). White arrows: NSP6-zippered domains of the NE, black arrows: regular NE. Insets, magnification of boxed areas showing regular NE (arrowheads: nuclear pore). g, h. EM showing regular (black arrows) and zippered (white arrows) NE domains in cells expressing HA-NSP6 treated with DMSO (g) or K22 (h). i, A cell not expressing NSP6 treated with K22 shows regular NE (black arrows). Insets in g–i, magnification of boxed areas showing regular NE (arrowheads: nuclear pores). j, Morphometric analysis of zippered NE surface in control and K22-treated cells expressing HA-NSP6. Single values are plotted, Medians are indicated, n = 20 cells, two-tailed unpaired t-test. Scale bars, a–c, 10 µm; d, 7.5 µm; e, 3.8 µm; f, 750 nm, inset 200 nm; g–i, 1 µm, insets 200 nm.
Extended Data Fig. 5 NSP(ΔSGF) is more prone to homodimerization and/or oligomerization than the reference NSP6.
a, Fluoromicrographs of stably expressing Flag-NSP6 or Flag-NSP6(ΔSGF) cells induced with doxycycline at the indicated times. b, Levels of HA-NSP6 and HA-NSP6(ΔSGF) clones induced overnight with doxycycline analysed by western blot with anti-HA antibody. GAPDH was used as loading control. c, Doxycycline-induced HeLa clones expressing HA-NSP6 or HA-NSP6(ΔSGF), or parental (CTRL) cells, were radiolabelled for 1 h with 35S-methionine/cysteine and chased for the indicated times. Samples were immunoprecipitated and analysed by SDS–PAGE gel autoradiography (top panels) and by western blot (bottom panels). The estimated half-life for HA-NSP6 and HA-NSP6(ΔSGF) was 5 h. d, Western blot of supernatant (S) and pellet (P) of lysates (at increasing Triton-X-100/NP-40 concentrations) of cells expressing NSP6-Flag, Flag-NSP6, or Flag-NSP6(ΔSGF) (see Methods). Numbers indicate the fraction of protein in the supernatant. e, Cell lysates (input) of cells expressing GFP-NSP6 with HA-NSP6, or GFP-NSP6(ΔSGF) with HA-NSP6(ΔSGF), were immunoprecipitated (IP) with anti-HA antibody and analysed by western blot with anti-HA and anti-GFP antibodies. The graph shows co-IP efficiency of NSP6(ΔSGF) relative to NSP6, which was set as 1 (see Methods). Mean ± SEM, n = 3 samples examined over three independent experiments. Two-tailed unpaired t-test with Welch’s correction. f, Fluorescence Loss in Photobleaching (FLIP) analysis of cells expressing GFP-NSP6 or GFP-NSP6(ΔSGF). Left panels, before bleaching. Right panels, after bleaching. Dashed lines indicate the areas in which iterative bleaching was applied. Graph: quantitative analysis of FLIP (see Methods). Values are expressed as a percentage of time 0, Means ± SD, three independent experiments, n = 10–12 cells per experiment. The calculated FLIP half-life for GFP-NSP6 is 70.5 s ± 12.6 and for GFP-NSP6(ΔSGF) 103.8 s ± 27.6. g, FRAP analysis of GFP-NSP6(ΔSGF)-expressing cells treated with DMSO or K22 for 16 h after bleaching of the individual NSP6(ΔSGF)-compartments (boxed). The small panels are representative frames (from a total of 100) at different times (in seconds) after the bleach. h, Quantitative FRAP analysis of the experiment in (g). Fluorescence intensity is expressed as a percentage of the value measured at time 0 (normalized to 100%). Means ± SD, three independent experiments, n = 45 structures. i, Doxycycline-induced clone expressing Flag-NSP6(ΔSGF) treated with DMSO or K22 for 24 h. Number of NSP6 structures (middle panel) and cells with elongated NSP6 structures (right panel) induced by K22. The number indicates the percentage (Mean ± SD) of cells exhibiting the elongated structures. Single values are plotted, Means ± SEM are indicated, N = 3, n = 90, two-tailed unpaired t-test with Welch’s correction. j–l. Immuno-CLEM of cells expressing HA-NSP6(ΔSGF). j, Fluoromicrograph (anti-HA immunostaining) of the NSP6 compartment. k, Magnification of the box in (j) (inset) and IEM, in which arrows 1–4 indicate overlap of the fluorescent and immuno-gold signals in the zippered NSP6-positive-structures. l, magnification of the structure indicated by arrow 1. m, Fluoromicrographs of cells expressing Flag-NSP6/NSP7-V5 or Flag-NSP6(ΔSGF)/NSP7-V5 immunostained with anti-Flag (green), anti-V5 (white insets) and anti-GM130 antibodies (red insets). Bottom panels, merge of Flag-NSP6 and GM130. n, Western blot of cell lysates from non-transfected (NT) and HA-NSP6/NSP7-V5 or HA-NSP6(ΔSGF)/NSP7-V5 expressing HeLa cells. Western blot images are representative of three independent experiments. Scale bars, a, f, g, i, m, 10 µm; j, 3.7 µm; k, 480 nm; l, 250 nm.
Extended Data Fig. 6 NSP6 organizes the DMVs induced by NSP3–NSP4.
a, Fluoromicrographs of HeLa cells expressing YFP-Cb5 with HA-NSP3 (anti-HA immunostaining) or mCherry-NSP4. Insets, merge with YFP-Cb5. b, Fluoromicrographs of HeLa cells expressing HA-NSP3 and mCherry-NSP4. Insets, enlargement of boxed area. Arrowheads, NSP3/NSP4-positive structures. Dashed lines delineate cell boundaries. c, Western blot (WB) of total lysates from HeLa cells expressing HA-NSP3, mCherry-NSP4, Flag-NSP6, or GFP as indicated. Actin was used as loading control. d, IEM and e, routine EM of HeLa cells co-transfected with HA-NSP3 and mCherry-NSP4. Anti-HA labelling in (d) shows gold particles decorating DMVs, indicated by asterisks. Black arrows, ER. Inset, magnification of boxed area. White arrows in d and e show double membranes. The average DMV size is 92 ± 30 nm. f, Western blot of total lysates from HeLa cells expressing HA-NSP3, mCherry-NSP4 or Flag-NSP6 as indicated. Actin was used as loading control. g, Individual fluoromicrographs of a Calu-3 cell co-transfected with Flag-NSP6, HA-NSP3, and mCherry-NSP4. h, Length of DMV–ER tubular or zippered connections in NSP3/NSP4 or in NSP3/NSP4/NSP6-expressing cells, respectively. Single values are plotted. Medians are shown (n ≥ 14 connections), two-tailed unpaired t-test. i, Tomographic slice of a HeLa cell expressing HA-NSP3/mCherry-NSP4 or j, HA-NSP3/mCherry-NSP4/Flag-NSP6, showing DMV clusters with regular round DMVs (white arrows) and large and elongated DMVs (black arrows). k, Frequency histograms of DMV diameter measured from tomograms of cells expressing NSP3/NSP4 (average diameter 80.87 nm) or NSP3/NSP4/NSP6 (average diameter 67.50 nm). Non-parametric Kolmogorov-Smirnov (KS) test. n≥135 vesicles. l, Tomographic slice of HeLa cells transfected with HA-NSP3/mCherry-NSP4 or m, HA-NSP3/mCherry-NSP4/Flag-NSP6, with arrows indicating the edges of the DMV clusters. n, DMV densities were calculated in tomograms as the number of vesicles per µm3 in a volume occupied by a DMV cluster. n = 8 clusters; Single values are plotted, Medians are shown, two-tailed unpaired t-test. Scale bars, a, b, g, 10 µm; i, j, l, m, 180 nm.
Extended Data Fig. 7 K22 impairs the ability of NSP6 to organize the NSP3–NSP4 puncta and has anti-SARS-CoV-2 activity.
a, HeLa cells transfected with HA-NSP3 and mCherry-NSP4 for 5 h were further transfected or not with Flag-NSP6 and treated with DMSO or K22 (40 μM, 16 h) followed by immunostaining as indicated. b, Quantification of the number of NSP4 puncta/cell in (a). N = 3, n = 60. Single values are plotted. The median value is shown. One-way ANOVA test with Tukey’s post-hoc. ns, not significant. c, d, CLEM analysis of K22-treated cells. c, Fluoromicrograph of HeLa cell expressing HA-NSP3, mCherry-NSP4 and Myc-NSP6. Inset corresponds to the boxed area and shows NSP3/NSP4-positive structures (arrows) close to the NSP6 compartment (arrowhead). d, Overlap of fluorescent image (inset in c) with EM image. The NSP6 compartment corresponds to a circular zippered ER structure (arrowhead) close to but not connected with the NSP3/NSP4 puncta that correspond to DMVs (arrows). The empty arrow indicates a tubular connection of a DMV to the regular ER (magnified in the inset). e, Ultrastructure of DMV clusters in K22-treated cells expressing HA-NSP3, mCherry-NSP4 and Myc-NSP6. Serial sections show a DMV cluster with irregular elongated DMVs (black arrows). The empty arrow indicates a tubular connection of a DMV with regular ER. f, Morphometric analysis of serial sections from untreated (NT) and K22-treated cells to quantify the number of DMVs per cluster, DMV shape factor, and the number of tubular or zippered connections per DMV cluster. Single values are plotted, Medians are shown, n = at least 8 clusters or 70 DMVs, two-tailed unpaired t-test. g, h, Antiviral activity of K22. Effects of K22 on cell death measured by LDH (g) or on viral replication (h) in SARS-CoV-2-infected Calu-3 cells. Mean ± SEM, N = 3, one-way ANOVA. i, Calu-3 cells infected with SARS-CoV-2 without (left panels) or with (right panel) K22 treatment. Cells were immunostained for dsRNA and NSP6. Nuclei were stained with DAPI. An enlargement of the boxed area shows NSP6 labelling (arrowheads) in the proximity of replication areas labelled by dsRNA. Graph, NSP6 structures within a distance of 250 nm from dsRNA spots, and dsRNA spots within a distance of 250 nm from NSP6 structures were counted and expressed as a percentage of total. Mean ± SEM, 15 cells analysed, n = 729 NSP6 structures, n = 901 dsRNA spots. Scale bar, a, 10 μm; c, 4.4 µm; d, 370 nm; e, 320 nm; i, 20 μm.
Extended Data Fig. 8 NSP6(ΔSGF) is more proficient in zippering the ER.
a, Tomographic slice of a HeLa cell expressing HA-NSP3/mCherry-NSP4/Flag-NSP6 or b, expressing HA-NSP3/mCherry-NSP4/Flag-NSP6(ΔSGF). Arrows indicate zippered ER connectors directed towards DMV clusters. c, Quantification of the number of ER zippered connections per DMV cluster and d, number of DMVs per cluster. Single values are plotted, Medians are shown. NSP3/NSP4/NSP6, N = 7 cells, n = 8 DMV clusters; NSP3/NSP4/NSP6(ΔSGF), N = 8 cells, n = 9 DMV clusters. Two-tailed unpaired t-test. e, Frequency histograms of DMV diameter measured from tomograms of cells expressing NSP3/NSP4/NSP6 (average diameter 67.50 nm) or NSP3/NSP4/NSP6(ΔSGF) (average diameter 68.51 nm). The histograms were analysed using non-parametric Kolmogorov-Smirnov (KS) test. n ≥ 123 vesicles. f, Serial tomographic slices from Calu-3 cells infected with 10 MOIs of an early lineage SARS-CoV-2 for 24 h. White arrows indicate a zippered connector that links ER (black arrows) to two DMVs (1 and 2). g, h, Serial tomographic slices from Calu-3 cells infected with the Gamma variant of SARS-CoV-2. White arrows indicate zippered connectors that depart from the ER (black arrows) and then branch (red arrow) towards two DMVs (1 and 2) in (g) and link the ER to a DMV (1) in h. White arrowheads indicate connectors that link DMV2 to DMV3 in (g) and DMV1 to DMV3 in (h). Scale bars, a, b 140 nm; f–h 200 nm.
Extended Data Fig. 9 RAB18 is recruited to ROLS by NSP6.
a, Fluoromicrographs of HeLa cells expressing Flag-NSP6-C80 (i.e. the last 80 amino acids of NSP6. Anti-Flag antibody and staining for LDs using BODIPY C12. Insets, enlargement of boxed areas. Arrowheads, colocalizing structures. b, Flag-NSP6-C80 associates with roundish structures that stain with BODIPY-488 and with ADRP (perilipin 2). c, NSP6-C80 mutated in residues that abrogate the amphiphilic properties of the AH fail to associate with LDs. Cells were immunostained with anti-Flag Ab (green) and with BODIPY-DA-C12 (red). d, Calu-3 cells infected with SARS-CoV-2 stained for LDs (Oil Red O staining)22 and immunostained with anti-dsRNA and anti-NSP6 antibodies. Blue, nuclear DAPI staining. Graph, quantification of the association of NSP6 or dsRNA positive structures with LDs, expressed as a percentage of the total number of NSP6 or dsRNA structures per cell. Mean ± SEM, N = 12, n = 1,377 and n = 861 for dsRNA and NSP6, respectively. e, Calu-3 cells infected or not with SARS-CoV-2, with or without the DGAT-1 inhibitor A922500, were analysed for LDs using Oil Red O staining (measurement of the fluorescent area of LDs- left graph) or for viral titres22 (right graph). Two-tailed unpaired t-test with Welch’s correction (left graph); one-way ANOVA followed by Tukey’s post-hoc test (right graph). f, Graph obtained applying the function Shuffle (see Methods) for the “NSP6 objects” and “LD objects” in the NSP3/NSP4/NSP6 transfected cell in Fig. 4a, showing that the measured distances (blue line) are significantly different from mean random distances (red line) flanked by 95% confidence intervals (green lines). g, Fluoromicrographs of Calu-3 cells expressing mCherry-NSP4, HA-NSP3, and Flag-NSP6 (left panel), or mCherry-NSP4 and HA-NSP3 (middle panel), or Flag-NSP6 (right panel). Cells were immunostained with anti-Flag Ab (blue). mCherry fluorescence was used as a read-out for mCherry-NSP4/HA-NSP3 structures. LDs were detected using BODIPY-488 (green). Inset, enlargement of boxed area. Arrowheads indicate LDs close to the NSP6 compartment and in proximity to the replication organelle. h, Cells expressing GFP-RAB18 alone or with Flag-NSP6 (anti-Flag immunostaining). Insets, arrowheads show co-localization of RAB18 with the NSP6 compartment. i, Individual fluoromicrographs and merge of cells co-expressing GFP-DFCP1, mCherry-NSP4 and HA-NSP3 (anti-HA immunostaining). j, In vitro pull-down assays using total cell lysates (input) from HeLa cells expressing HA-NSP6 incubated with GST-DFCP1 or GST alone. Upper panel, Ponceau Red staining; bottom panel, western blot with anti-HA antibody. Images are representative of three independent experiments. Scale bars, a–c, g–i, 10 μm; d, 20 μm.
Extended Data Fig. 10 The C-terminal domain of NSP6 is involved in the recruitment of DFCP1 in a PtdIns3P-independent manner.
a, Fluoromicrographs of cells co-expressing GFP-DFCP1 and Flag-NSP6(1–157). b, Fluorescent images of HeLa cells transfected with the indicated DFCP1 mutants alone or in combination with NSP6 (as indicated). A schematic representation of DFCP1 mutants is reported on top. Arrowheads, DFCP1 signal in the NSP6 compartment. Insets, enlarged merge of boxed areas. Numbers indicate the percentage of co-localization between DFCP1 mutants and NSP6. Mean ± SD (see Methods). c, Fluorescent micrographs of cells expressing mCherry-DFCP1 and Flag-NSP6 treated with SAR405 or with wortmannin. Anti-Flag immunostaining. d, Fluoromicrograph showing LC3 staining in one transfected and one non-transfected cell from Flag-NSP6-expressing HeLa cells. Inset, anti-Flag immunostaining. Graph, quantification of LC3 puncta in non-transfected (CTRL) and NSP6-transfected cells (number of LC3 spots/cell). Means ± SEM. n = 74 cells examined over three independent experiments. Two-tailed unpaired t-test with Welch’s correction. ns, not significant. e, HeLa cells expressing HA-NSP3 and GFP-NSP4, or HA-NSP3, GFP-NSP4 and Flag-NSP6, were loaded with BODIPY-DA-C12 and washed out for 6 h (see Methods). Dotted yellow lines, LDs; dotted white lines, NSP4 puncta. Graph, quantification of the percentage of NSP4 puncta positive for BODIPY-DA-C12. Single values are plotted, Means ± SEM are indicated, N = 3, n = 90, two-tailed Mann–Whitney test. f, Western blot of protein extracts from mock-treated (CTRL) and DFCP1-KD cells. Actin was used as loading control. The graph shows the level of DFCP1 protein expressed as percentage of control (set at 100). Mean ± SEM. N = 3, Two-tailed unpaired t-test with Welch’s correction. g, Western blot of protein lysates from untreated (CTRL), scramble- and DFCP1 siRNA-treated (DFCP1-KD) SARS-CoV-2-infected Calu-3 cells detected with an anti-DFCP1 antibody. GAPDH was used as loading control. The graph shows the level of DFCP1-KD expressed as percentage of control (set at 100). Mean ± SEM. N = 3, two-tailed unpaired t-test with Welch’s correction. h, Quantification of viral titres in SARS-CoV-2-infected Calu-3 cells untreated (-), transfected with scramble siRNA (CTRL) or DFCP1 siRNA (DFCP1-KD). Mean ± SEM, N = 7, two-tailed unpaired t-test with Welch’s correction. Scale bar a–e, 10 μm. Western blot images are representative of three independent experiments.
Extended Data Fig. 11 Working model for the role of NSP6 in replication organelle biogenesis.
NSP6-induced zippered connectors are cues and organizers for NSP3/NSP4-induced DMV formation acting as selective communication tracks with the ER (largely excluding luminal ER proteins). In addition, the connectors might also act as fast-tracks to refurbish the actively growing subpopulation of DMVs with lipids derived from LDs.
Supplementary information
Supplementary Information
This file contains legends for Supplementary Videos 1–14, Supplementary Tables 1–2 and Supplementary Figure 1.
Video 1 Electron tomogram of HeLa cells transfected with HA–NSP6.
Connections of circular zippered structures with the ER are shown by black arrows, and the connection of linear zippered membranes with the nuclear envelope is indicated by a white arrow.
Video 2 FRAP experiment of HeLa cells transfected with GFP–VAP-A and mCherry–NSP6.
The bleached region is highlighted in the boxed area.
Video 3 FRAP experiment of HeLa cells transfected with YFP–Cb5 and mCherry–NSP6.
The bleached region is highlighted in the boxed area.
Video 4 FRAP experiment of HeLa cells transfected with mCherry–NSP6 and incubated with BODIPY C12-HPC.
The bleached region is highlighted in the boxed area.
Video 5 Electron tomogram of HeLa cells transfected with HA–NSP3, mCherry–NSP4, and GFP–NSP6.
White arrows show connections of zippered ER to DVMs. Black arrows show connection of zippered region to regular ER cisternae.
Video 6 3D reconstruction of the tomogram shown in EM Video 5.
DMVs, zippered ER, and regular ER are shown in red, green and yellow, respectively.
Video 7 Electron tomogram of HeLa cells transfected with HA–NSP3, mCherry–NSP4, and GFP–NSP6.
Another example from electron tomogram of connections between zippered ER and DVMs (white arrows) and between zippered ER and regular ER cisternae (black arrow).
Video 8 Electron tomogram of HeLa cells transfected with HA–NSP3 and mCherry–NSP4.
White arrows show connections between ER and DVMs.
Video 9 3D reconstruction of the tomogram shown in EM Video 8.
DMVs and ER are shown in red and yellow, respectively.
Video 10 Electron tomogram of HeLa cells transfected with HA–NSP3 and mCherry–NSP4.
Another example from electron tomogram of connections between ER and DVMs (white arrows).
Video 11 Electron tomogram of HeLa cells transfected with HA–NSP3, mCherry–NSP4, and GFP–NSP6(ΔSGF).
White arrows show numerous connections of zippered ER to DVMs. Black arrow shows connection of zippered region to regular ER cisternae.
Video 12 3D reconstruction of the tomogram shown in EM Video 11.
DMVs, zippered ER, and regular ER are shown in red, green and yellow, respectively.
Video 13 Electron tomogram of Calu-3 cells infected with early lineage B.1 SARS-CoV-2.
White arrow shows zippered DMV connector.
Video 14 Electron tomogram of Calu-3 cells infected with the Gamma variant of SARS-CoV-2.
White arrows show long zippered ER connectors directed to DMVs.
Source data
Rights and permissions
About this article
Cite this article
Ricciardi, S., Guarino, A.M., Giaquinto, L. et al. The role of NSP6 in the biogenesis of the SARS-CoV-2 replication organelle. Nature 606, 761–768 (2022). https://doi.org/10.1038/s41586-022-04835-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-04835-6
This article is cited by
-
Lipid droplets in pathogen infection and host immunity
Acta Pharmacologica Sinica (2024)
-
SARS-CoV-2 biology and host interactions
Nature Reviews Microbiology (2024)
-
Innate immune evasion strategies of SARS-CoV-2
Nature Reviews Microbiology (2023)
-
SARS-CoV-2 variant biology: immune escape, transmission and fitness
Nature Reviews Microbiology (2023)
-
SARS-CoV-2 nsp3 and nsp4 are minimal constituents of a pore spanning replication organelle
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.