Abstract
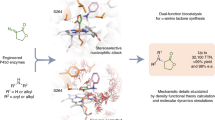
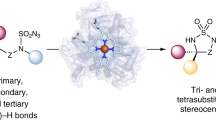
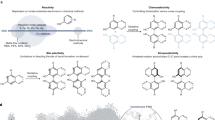
Chiral amine diastereomers are ubiquitous in pharmaceuticals and agrochemicals1, yet their preparation often relies on low-efficiency multi-step synthesis2. These valuable compounds must be manufactured asymmetrically, as their biochemical properties can differ based on the chirality of the molecule. Herein we characterize a multifunctional biocatalyst for amine synthesis, which operates using a mechanism that is, to our knowledge, previously unreported. This enzyme (EneIRED), identified within a metagenomic imine reductase (IRED) collection3 and originating from an unclassified Pseudomonas species, possesses an unusual active site architecture that facilitates amine-activated conjugate alkene reduction followed by reductive amination. This enzyme can couple a broad selection of α,β-unsaturated carbonyls with amines for the efficient preparation of chiral amine diastereomers bearing up to three stereocentres. Mechanistic and structural studies have been carried out to delineate the order of individual steps catalysed by EneIRED, which have led to a proposal for the overall catalytic cycle. This work shows that the IRED family can serve as a platform for facilitating the discovery of further enzymatic activities for application in synthetic biology and organic synthesis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data supporting the findings of this study are available within the paper and its Supplementary Information, and NMR traces are available from the Mendeley data repository (https://data.mendeley.com) at https://doi.org/10.17632/fhc429t33c.1. Sequence data have been deposited in Genbank (accession numbers MW854365, MW925135–MW925140) and the coordinate files and structure factors have been deposited in the PDB with accession number 7A3W.
References
Jarvis, L. M. The new drugs of 2019. Chem. Eng. News 98, 30–36 (2020).
Afanasyev, O. I., Kuchuk, E., Usanov, D. L. & Chusov, D. Reductive amination in the synthesis of pharmaceuticals. Chem. Rev. 119, 11857–11911 (2019).
Marshall, J. R. et al. Screening and characterization of a diverse panel of metagenomic imine reductases for biocatalytic reductive amination. Nat. Chem. 13, 140–148 (2021).
Roughley, S. D. & Jordan, A. M. The medicinal chemist’s toolbox: an analysis of reactions used in the pursuit of drug candidates. J. Med. Chem. 54, 3451–3479 (2011).
Yasukawa, T., Masuda, R. & Kobayashi, S. Development of heterogeneous catalyst systems for the continuous synthesis of chiral amines via asymmetric hydrogenation. Nat. Catal. 2, 1088–1092 (2019).
Wu, Z. et al. Secondary amines as coupling partners in direct catalytic asymmetric reductive amination. Chem. Sci. 10, 4509–4514 (2019).
Skrypai, V., Varjosaari, S. E., Azam, F., Gilbert, T. M. & Adler, M. J. Chiral Brønsted acid-catalyzed metal-free asymmetric direct reductive amination using 1-hydrosilatrane. J. Org. Chem. 84, 5021–5026 (2019).
Aleku, G. A. et al. A reductive aminase from Aspergillus oryzae. Nat. Chem. 9, 961–969 (2017).
Mayol, O. et al. A family of native amine dehydrogenases for the asymmetric reductive amination of ketones. Nat. Catal. 2, 324–333 (2019).
Yang, Y., Cho, I., Qi, X., Liu, P. & Arnold, F. H. An enzymatic platform for the asymmetric amination of primary, secondary and tertiary C(sp3)–H bonds. Nat. Chem. 11, 987–993 (2019).
Li, T. et al. Efficient, chemoenzymatic process for, anufacture of the Boceprevir bicyclic [3.1.0]proline intermediate based on amine oxidase-catalyzed desymmetrization.J. Am. Chem. Soc. 134, 6467–6472 (2012).
Zhou, J. & List, B. Organocatalytic asymmetric reaction cascade to substituted cyclohexylamines. J. Am. Chem. Soc. 129, 7498–7499 (2007).
Monti, D. et al. Cascade coupling of ene-reductases and ω-transaminases for the stereoselective synthesis of diastereomerically enriched amines. ChemCatChem 7, 3106–3109 (2015).
France, S. P., Hepworth, L. J., Turner, N. J. & Flitsch, S. L. Constructing biocatalytic cascades: in vitro and in vivo approaches to de novo multi-enzyme pathways. ACS Catal. 7, 710–724 (2017).
Huffman, M. A. et al. Design of an in vitro biocatalytic cascade for the manufacture of islatravir. Science 366, 1255–1259 (2019).
Toogood, H. S. & Scrutton, N. S. Discovery, Characterization, engineering, and applications of ene-reductases for industrial biocatalysis. ACS Catal. 8, 3532–3549 (2018).
Roth, S., Kilgore, M. B., Kutchan, T. M. & Müller, M. Exploiting the catalytic diversity of short-chain dehydrogenases/reductases: versatile enzymes from plants with extended imine substrate scope. ChemBioChem 19, 1849–1852 (2018).
Hyslop, J. F. et al. Biocatalytic synthesis of chiral N-functionalized amino acids. Angew. Chemie. Int. Ed. 57, 13821–13824 (2018).
Kato, Y., Yamada, H. & Asano, Y. Stereoselective synthesis of opine-type secondary amine carboxylic acids by a new enzyme opine dehydrogenase use of recombinant enzymes. J. Mol. Catal. B 1, 151–160 (1996).
Schober, M. et al. Chiral synthesis of LSD1 inhibitor GSK2879552 enabled by directed evolution of an imine reductase. Nat. Catal. 2, 909–915 (2019).
Bornadel, A. et al. Technical considerations for scale-up of imine-reductase-catalyzed reductive amination: a case study. Org. Process Res. Dev. 23, 1262–1268 (2019).
Hussain, S. et al. An (R)-imine reductase biocatalyst for the asymmetric reduction of cyclic imines. ChemCatChem 7, 579–583 (2015).
Yao, P., Xu, Z., Yu, S., Wu, Q. & Zhu, D. Imine reductase-catalyzed enantioselective reduction of bulky α,β-unsaturated imines en route to a pharmaceutically important morphinan skeleton. Adv. Synth. Catal. 361, 556–561 (2019).
Mitsukura, K. et al. Purification and characterization of a novel (R)-imine reductase from Streptomyces sp. GF3587. Biosci. Biotechnol. Biochem. 75, 1778–1782 (2011).
Lenz, M. et al. Asymmetric ketone reduction by imine reductases. ChemBioChem 18, 253–256 (2017).
Thorpe, T. W. et al. One-pot biocatalytic cascade reduction of cyclic enimines for the preparation of diastereomerically enriched N-heterocycles. J. Am. Chem. Soc. 141, 19208–19213 (2019).
Steiningerova, L. et al. Different reaction specificities of F420H2-dependent reductases facilitate pyrrolobenzodiazepines and lincomycin to fit their biological targets. J. Am. Chem. Soc. 142, 3440–3448 (2020).
Trenti, F. et al. Early and late steps of quinine biosynthesis. Org. Lett. 23, 1793–1797 (2021).
Mitsukura, K., Suzuki, M., Tada, K., Yoshida, T. & Nagasawa, T. Asymmetric synthesis of chiral cyclic amine from cyclic imine by bacterial whole-cell catalyst of enantioselective imine reductase. Org. Biomol. Chem. 8, 4533–4535 (2010).
France, S. P. et al. Identification of novel bacterial members of the imine reductase enzyme family that perform reductive amination. ChemCatChem 10, 510–514 (2018).
Mangas-Sanchez, J. et al. Asymmetric synthesis of primary amines catalyzed by thermotolerant fungal reductive aminases. Chem. Sci. 11, 5052–5057 (2020).
Montgomery, S. L. et al. Characterization of imine reductases in reductive amination for the exploration of structure–activity relationships. Sci. Adv. 6, 9320 (2020).
Ouellet, S. G., Walji, A. M. & Macmillan, D. W. C. Enantioselective organocatalytic transfer hydrogenation reactions using Hantzsch esters. Acc. Chem. Res. 40, 1327–1339 (2007).
Santi, N., Morrill, L. C., Świderek, K., Moliner, V. & Luk, L. Y. P. Transfer hydrogenations catalyzed by streptavidin-hosted secondary amine organocatalysts. Chem. Commun. 57, 1919–1922 (2021).
Rodríguez-Mata, M. et al. Structure and activity of NADPH-dependent reductase Q1EQE0 from Streptomyces kanamyceticus, which catalyses the R-selective reduction of an imine substrate. ChemBioChem 14, 1372–1379 (2013).
Holm, L. Benchmarking fold detection by DaliLite v.5. Bioinformatics 35, 5326–5327 (2019).
Lenz, M. et al. New imine-reducing enzymes from β-hydroxyacid dehydrogenases by single amino acid substitutions. Protein Eng. Des. Sel. 31, 109–120 (2018).
Huber, T. et al. Direct reductive amination of ketones: structure and activity of S-selective imine reductases from Streptomyces. ChemCatChem 6, 2248–2252 (2014).
Man, H. et al. Structure, activity and stereoselectivity of NADPH-dependent oxidoreductases catalysing the S-selective reduction of the imine substrate 2-methylpyrroline. ChemBioChem 16, 1052–1059 (2015).
Aleku, G. A. et al. Stereoselectivity and structural characterization of an imine reductase (IRED) from Amycolatopsis orientalis. ACS Catal. 6, 3880–3889 (2016).
Trott, O. & Olson, A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2009).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296 (2021).
Kabsch, W. XDS. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 125–132 (2010).
Evans, P. Scaling and assessment of data quality. Acta Crystallogr. Sect. D Biol. Crystallogr. 62, 72–82 (2006).
Winter, G. xia2: an expert system for macromolecular crystallography data reduction. J. Appl. Crystallogr. 43, 186–190 (2010).
Vagin, A. & Teplyakov, A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 (1997).
Sharma, M. et al. A mechanism for reductive amination catalyzed by fungal reductive aminases. ACS Catal. 8, 11534–11541 (2018).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997).
Acknowledgements
T.W.T. is grateful for a CASE award from the UK Biotechnology and Biological Sciences Research Council (BBSRC) and Pfizer (BB/M011208/1). J.R.M. acknowledges a CASE award from the Industrial Biotechnology Innovation Centre (IBioIC), BBSRC and Prozomix Ltd. A.C. was funded by grant BB/P005578/1 from the BBSRC. N.J.T. is grateful to the ERC for the award of an Advanced Grant (742987). We thank J. P. Turkenburg and S. Hart for assistance with X-ray data collection and the Diamond Light Source for access to beamline I03 under proposal number mx-9948.
Author information
Authors and Affiliations
Contributions
N.J.T., G.G., R.M.H., R.K. and D.S.B.D. devised and supervised the project. F.P. and R.E.R. managed the project. T.W.T. and A.A. performed mechanistic studies. T.W.T. and R.E.R. carried out substrate scope reactions. T.W.T. and V.H. carried out preparative scale reactions. T.W.T., R.E.R and V.H. synthesized substrates and standards. A.C., T.W.T. and G.G. performed crystallographic and docking studies. T.W.T. and R.S.H. undertook site-directed mutagenesis. J.R.M., S.J.C. and J.D.F. performed genetic identification, cloning and bioinformatics. T.W.T. and J.R.M. produced and purified the biocatalyst. N.J.T., G.G., R.M.H., R.K., D.S.B.D., S.J.C., F.P., J.D.F., A.C., R.E.R., V.H., J.R.M. and T.W.T. wrote the manuscript and generated the figures.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Dominic Campopiano, Sandy Schmidt and Thomas Ward for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Phylogenetic IRED tree mapped against the reaction profiles of IRED-catalysed reduction of ene-imine I.
Although the majority of the IREDs catalysed conventional imine reduction only, a small number were able to reduce both C=C and C=N bonds. Of these, EneIRED (pIR-120) possessed the highest propensity to forming the desired amine II.
Extended Data Fig. 2 Optimization of EneIRED-catalysed CR–RA reaction conditions.
Conversion to the products of CR and CR–RA were elevated in glycine-OH pH 9.0, at moderate DMSO cosolvent concentration and at higher equivalents of amine donor. Formation of the direct RA product was not observed under any conditions.
Extended Data Fig. 3 Scaled-up examples of EneIRED-catalysed CR–RA.
Several secondary and tertiary amines could be prepared including an example at elevated enone concentration and lower amine equivalents.
Extended Data Fig. 4 Control reactions and isolated reactions of potential CR–RA pathway intermediates in EneIRED-catalysed CR–RA.
a, EneIRED CR–RA of 15 and b with NADPH. b, No enzyme control reaction. c, No recycling system control reaction. d, Reactions of potential CR–RA intermediate 15b with NADP+ or NADPH. e, Reaction of potential CR–RA intermediate 15′ with b using EneIRED. f, No amine control reactions with EneIRED point variants.
Extended Data Fig. 5 Time-course studies of the CR–RA of 15 with b catalysed by wild-type EneIRED and point variants.
Both EneIRED-Y177A and EneIRED-Y181A exhibited a reduction in the rate of CR and CR–RA product formation compared to wild-type EneIRED, indicating that both residues are important for efficient catalysis. Notably, for EneIRED-Y177A the concentration of the ketone intermediate was comparatively low throughout the reaction, suggesting that Y177 is more important for CR than RA.
Extended Data Fig. 6 Active site of EneIRED highlighting electron density.
a, Side chains, with density corresponding to the refined 2Fo − Fc map (blue) at a level of 1σ. b, NADP+, with density corresponding to the Fo − Fc difference map (green) at a level of 3σ obtained from refinement in the absence of the ligand, with refined atoms included for clarity. Fo and Fc stand for the observed and calculated structure factor amplitudes, respectively.
Supplementary information
Supplementary Information
This file contains Supplementary Text, Supplementary Figures, Supplementary Discussion and Supplementary References
Rights and permissions
About this article
Cite this article
Thorpe, T.W., Marshall, J.R., Harawa, V. et al. Multifunctional biocatalyst for conjugate reduction and reductive amination. Nature 604, 86–91 (2022). https://doi.org/10.1038/s41586-022-04458-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-04458-x
This article is cited by
-
Transaminase-catalysis to produce trans-4-substituted cyclohexane-1-amines including a key intermediate towards cariprazine
Communications Chemistry (2024)
-
Facile access to C-N bonds via unexpected side reactions of Knoevenagel condensation and their application in high-efficiency organic solar cells
Science China Chemistry (2024)
-
Active and stable alcohol dehydrogenase-assembled hydrogels via synergistic bridging of triazoles and metal ions
Nature Communications (2023)
-
Enantioconvergent Cu-catalysed N-alkylation of aliphatic amines
Nature (2023)
-
Buy one, get one free
Nature Synthesis (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.