Abstract
Ribosome rescue pathways recycle stalled ribosomes and target problematic mRNAs and aborted proteins for degradation1,2. In bacteria, it remains unclear how rescue pathways distinguish ribosomes stalled in the middle of a transcript from actively translating ribosomes3,4,5,6. Here, using a genetic screen in Escherichia coli, we discovered a new rescue factor that has endonuclease activity. SmrB cleaves mRNAs upstream of stalled ribosomes, allowing the ribosome rescue factor tmRNA (which acts on truncated mRNAs3) to rescue upstream ribosomes. SmrB is recruited to ribosomes and is activated by collisions. Cryo-electron microscopy structures of collided disomes from E. coli and Bacillus subtilis show distinct and conserved arrangements of individual ribosomes and the composite SmrB-binding site. These findings reveal the underlying mechanisms by which ribosome collisions trigger ribosome rescue in bacteria.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Cryo-EM volumes and molecular models have been deposited at the Electron Microscopy Data Bank (EMDB) and Protein Data Bank (PDB) with the following respective accession codes: for the E. coli disome, EMD-13952 and 7QG8 (stalled 70S) and EMD-13955 and 7QGH (collided 70S); for the E. coli trisome, EMD-13964; for the B. subtilis disome, EMD-13959 and 7QGU (stalled 70S) and EMD-13961 and 7QH4 (collided 70S); for the E. coli disome–SmrB complex, EMD-13956 and 7QGN (stalled 70S) and EMD-13958 and 7QGR (collided 70S). Gel source images are provided in Supplementary Figs. 1 and 2.
Code availability
Custom Python scripts used to analyse the Tn-seq and RACE data are freely available at https://github.com/greenlabjhmi/2021_SmrB.
References
Müller, C., Crowe-McAuliffe, C. & Wilson, D. N. Ribosome rescue pathways in bacteria. Front. Microbiol. 12, 652980 (2021).
Keiler, K. C., Waller, P. R. & Sauer, R. T. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271, 990–993 (1996).
Ivanova, N., Pavlov, M. Y., Felden, B. & Ehrenberg, M. Ribosome rescue by tmRNA requires truncated mRNAs. J. Mol. Biol. 338, 33–41 (2004).
Hayes, C. S. & Sauer, R. T. Cleavage of the A site mRNA codon during ribosome pausing provides a mechanism for translational quality control. Mol. Cell 12, 903–911 (2003).
Subramaniam, A. R., Zid, B. M. & O’Shea, E. K. An integrated approach reveals regulatory controls on bacterial translation elongation. Cell 159, 1200–1211 (2014).
Janssen, B. D., Garza-Sánchez, F. & Hayes, C. S. A-site mRNA cleavage is not required for tmRNA-mediated ssrA-peptide tagging. PLoS ONE 8, e81319 (2013).
Moore, S. D. & Sauer, R. T. Ribosome rescue: tmRNA tagging activity and capacity in Escherichia coli. Mol. Microbiol. 58, 456–466 (2005).
Yan, L. L. & Zaher, H. S. How do cells cope with RNA damage and its consequences? J. Biol. Chem. 294, 15158–15171 (2019).
Thomas, E. N., Kim, K. Q., McHugh, E. P., Marcinkiewicz, T. & Zaher, H. S. Alkylative damage of mRNA leads to ribosome stalling and rescue by trans translation in bacteria. eLife 9, e61984 (2020).
Roche, E. D. & Sauer, R. T. SsrA-mediated peptide tagging caused by rare codons and tRNA scarcity. EMBO J. 18, 4579–4589 (1999).
Sunohara, T., Jojima, K., Tagami, H., Inada, T. & Aiba, H. Ribosome stalling during translation elongation induces cleavage of mRNA being translated in Escherichia coli. J. Biol. Chem. 279, 15368–15375 (2004).
Hayes, C. S., Bose, B. & Sauer, R. T. Proline residues at the C terminus of nascent chains induce SsrA tagging during translation termination. J. Biol. Chem. 277, 33825–33832 (2002).
Neubauer, C., Gillet, R., Kelley, A. C. & Ramakrishnan, V. Decoding in the absence of a codon by tmRNA and SmpB in the ribosome. Science 335, 1366–1369 (2012).
Ikeuchi, K. et al. Collided ribosomes form a unique structural interface to induce Hel2-driven quality control pathways. EMBO J. 38, e100276 (2019).
Juszkiewicz, S. et al. ZNF598 is a quality control sensor of collided ribosomes. Mol. Cell 72, 469–481 (2018).
Matsuo, Y. et al. Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nat. Commun. 8, 159 (2017).
Simms, C. L., Yan, L. L. & Zaher, H. S. Ribosome collision is critical for quality control during no-go decay. Mol. Cell 68, 361–373 (2017).
Langridge, G. C. et al. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res. 19, 2308–2316 (2009).
Kannan, K. et al. The general mode of translation inhibition by macrolide antibiotics. Proc. Natl Acad. Sci. USA 111, 15958–15963 (2014).
Beckert, B. et al. Structural and mechanistic basis for translation inhibition by macrolide and ketolide antibiotics. Nat. Commun. 12, 4466 (2021).
Zhou, W. et al. PPR-SMR protein SOT1 has RNA endonuclease activity. Proc. Natl Acad. Sci. USA 114, E1554–E1563 (2017).
Wu, W. J. et al. SOT1, a pentatricopeptide repeat protein with a small MutS-related domain, is required for correct processing of plastid 23S-4.5S rRNA precursors in Arabidopsis thaliana. Plant J. 85, 607–621 (2016).
D’Orazio, K. N. et al. The endonuclease Cue2 cleaves mRNAs at stalled ribosomes during no go decay. eLife 8, e49117 (2019).
Liu, S., Melonek, J., Boykin, L. M., Small, I. & Howell, K. A. PPR-SMRs: ancient proteins with enigmatic functions. RNA Biol. 10, 1501–1510 (2013).
Glover, M. L. et al. NONU-1 encodes a conserved endonuclease required for mRNA translation surveillance. Cell Rep. 30, 4321–4331 (2020).
Mohammad, F., Green, R. & Buskirk, A. R. A systematically-revised ribosome profiling method for bacteria reveals pauses at single-codon resolution. eLife 8, e42591 (2019).
Chiba, S., Lamsa, A. & Pogliano, K. A ribosome-nascent chain sensor of membrane protein biogenesis in Bacillus subtilis. EMBO J. 28, 3461–3475 (2009).
Ishii, E. et al. Nascent chain-monitored remodeling of the Sec machinery for salinity adaptation of marine bacteria. Proc. Natl Acad. Sci. USA 112, E5513–E5522 (2015).
Su, T. et al. The force-sensing peptide VemP employs extreme compaction and secondary structure formation to induce ribosomal stalling. eLife 6, e25642 (2017).
Schuwirth, B. S. et al. Structures of the bacterial ribosome at 3.5 Å resolution. Science 310, 827–834 (2005).
Selmer, M., Gao, Y. G., Weixlbaumer, A. & Ramakrishnan, V. Ribosome engineering to promote new crystal forms. Acta Crystallogr. D 68, 578–583 (2012).
Atkins, J. F. & Bjork, G. R. A gripping tale of ribosomal frameshifting: extragenic suppressors of frameshift mutations spotlight P-site realignment. Microbiol. Mol. Biol. R. 73, 178 (2009).
Beckert, B. et al. Structure of the Bacillus subtilis hibernating 100S ribosome reveals the basis for 70S dimerization. EMBO J. 36, 2061–2072 (2017).
Beckert, B. et al. Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1. Nat. Microbiol. 3, 1115 (2018).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Ferrin, M. A. & Subramaniam, A. R. Kinetic modeling predicts a stimulatory role for ribosome collisions at elongation stall sites in bacteria. eLife 6, e23629 (2017).
Simms, C. L., Yan, L. L., Qiu, J. K. & Zaher, H. S. Ribosome collisions result in +1 frameshifting in the absence of no-go decay. Cell Rep. 28, 1679–1689 (2019).
Smith, A. M., Costello, M. S., Kettring, A. H., Wingo, R. J. & Moore, S. D. Ribosome collisions alter frameshifting at translational reprogramming motifs in bacterial mRNAs. Proc. Natl Acad. Sci. USA 116, 21769–21779 (2019).
Chai, Q. et al. Organization of ribosomes and nucleoids in Escherichia coli cells during growth and in quiescence. J. Biol. Chem. 289, 11342–11352 (2014).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645 (2000).
Crameri, A., Whitehorn, E. A., Tate, E. & Stemmer, W. P. C. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat. Biotechnol. 14, 315–319 (1996).
Jiang, H. S., Lei, R., Ding, S. W. & Zhu, S. F. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 15, 182 (2014).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Eddy, S. R. A new generation of homology search tools based on probabilistic inference. Genome Inform. 23, 205–211 (2009).
Finn, R. D. et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285 (2016).
Lassmann, T., Frings, O. & Sonnhammer, E. L. L. Kalign2: high-performance multiple alignment of protein and nucleotide sequences allowing external features. Nucleic Acids Res. 37, 858–865 (2009).
Cole, C., Barber, J. D. & Barton, G. J. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36, W197–W201 (2008).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490 (2010).
Shevchenko, A., Wilm, M., Vorm, O. & Mann, M. Mass spectrometric sequencing of proteins from silver stained polyacrylamide gels. Anal. Chem. 68, 850–858 (1996).
Sohmen, D. et al. Structure of the Bacillus subtilis 70S ribosome reveals the basis for species-specific stalling. Nat. Commun. 6, 6941 (2015).
Schafer, H. et al. The alarmones (p)ppGpp are part of the heat shock response of Bacillus subtilis. PLoS Genet. 16, e1008275 (2020).
Wells, J. N. et al. Structure and function of yeast Lso2 and human CCDC124 bound to hibernating ribosomes. PLoS Biol. 18, e3000780 (2020).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Zivanov, J., Nakane, T. & Scheres, S. H. W. Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ 7, 253–267 (2020).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Loveland, A. B., Demo, G. & Korostelev, A. A. Cryo-EM of elongating ribosome with EFTu-GTP elucidates tRNA proofreading. Nature 584, 640–645 (2020).
Byrne, R. T., Konevega, A. L., Rodnina, M. V. & Antson, A. A. The crystal structure of unmodified tRNAPhe from Escherichia coli. Nucleic Acids Res. 38, 4154–4162 (2010).
Loveland, A. B. & Korostelev, A. A. Structural dynamics of protein S1 on the 70S ribosome visualized by ensemble cryo-EM. Methods 137, 55–66 (2018).
Mirdita, M., Steinegger, M. & Soding, J. MMseqs2 desktop and local web server app for fast, interactive sequence searches. Bioinformatics 35, 2856–2858 (2019).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Acknowledgements
We thank S. Sanyal for sharing E. coli strains QC101 and QC901, H. Hao at the JHMI Transcriptomics and Deep Sequencing Core for providing assistance with high-throughput sequencing, B. Cole and T. Boronina at JHMI in the Mass Spectrometry and Proteomics Facility, J. Musial for providing assistance during protein purification, T. Mackens-Kiani for helping with the nuclease assay data analysis, C. Ungewickell and S. Rieder for providing technical assistance, and L. Kater and K. Best for offering support with the preprocessing pipeline for the cryo-EM data. This work was supported by NIH grant GM136960 (A.R.B.), HHMI (R.G.), the Intramural Research Program of the National Library of Medicine at the NIH (A.M.B. and L.A.) and the German Research Council (TRR174) (R. Beckmann). H.K. is supported by a DFG fellowship through the Graduate School of Quantitative Bioscience Munich (QBM).
Author information
Authors and Affiliations
Contributions
K.S. performed the genetic screening, analysis of the NanoLuc–ble reporters and the sucrose gradients. A.C. analysed the CRP reporters and prepared samples for the MS experiments. A.M.B. and L.A. performed the phylogenetic analyses. H.K. performed the in vitro translation and in vitro nuclease assays, prepared samples for cryo-EM analysis and processed the cryo-EM data. O.B. and R. Buschauer collected the cryo-EM data. H.K. and R. Buschauer prepared the molecular models. H.K., R. Buschauer and R. Beckmann analysed and interpreted the structures, and R. Buschauer prepared the structural figures. R.G., R. Beckmann and A.R.B. supervised the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Vasili Hauryliuk, Yury Polikanov and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 SmrB is a general ribosome rescue factor.
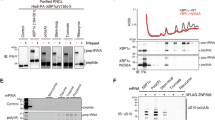
a, Reporter protein from wild-type and ∆ssrA strains was detected by antibodies against the N-terminal Strep-tag. Arrows indicate the full-length fusion protein (FL) and shorter NanoLuc protein (N). The RpoB protein serves as a loading control. b, Additional reporters to study ribosome rescue in E. coli with various stalling motifs. c, The expression of full-length NanoLuc-Ble protein was monitored with an anti-Flag antibody; anti-Strep antibodies reveal both full-length NanoLuc-Ble and truncated NanoLuc protein. RpoB serves as a loading control.
Extended Data Fig. 2 Phylogenetic tree of SMR domain proteins.
Stylized phylogenetic tree depicting relationships between SMR domain clades. Clades with indicated bootstrap support are marked with circles. Clade names are given to the right of the tree. Dotted lines indicate positions with little or no bootstrap support.
Extended Data Fig. 3 Distribution and architectures of SMR-domain proteins.
a, Heat map demonstrating the conservation and distribution of SMR-domain proteins and other related translational quality control factors. Smr-all includes all types of SMR-domain proteins; Smr-euk includes only the eukaryotic branch. b, Domain organization of three representative bacterial proteins containing an SMR domain. c, Multiple alignment of the conserved regions in the N-terminal extension of SMR proteins from proteobacteria. Columns in the alignment are shaded and labeled according to biochemical character: -, negatively charged; h, hydrophobic in yellow; a, aromatic; p, polar in blue; l, aliphatic in yellow; s, small in green; u, tiny in green. Residue positions in the–xxxa motif are colored in white and shaded in black, marked by asterisks above the alignment. Residue positions forming part of the active site of the core SMR domain are colored in white and shaded in red. Sequences are labeled with NCBI accession number and organism abbreviation; abbreviations are provided below alignment. Secondary structure provided at top of alignment. Numbers to left and right of alignment denote positioning of the region. Internal numbers give the size of excised variable insert regions. d, Sequence alignment of SMR domains of representative proteins. Identical residues are shown in white with a red background; conserved residues are shown in red. The identity of each sequence is represented by the gene name, species name, and numbers indicating the beginning and the end of the residues used for the alignment. Ecol, Escherichia coli; Scer, Saccharomyces cerevisiae; Cele, Caenorhabditis elegans; Hsap, Homo sapiens; Atha, Arabidopsis thaliana; Bsub, Bacillus subtilis.
Extended Data Fig. 4 SmrB cleavage, tmRNA tagging, and ribosome collisions.
a, The results of 5′-RACE showing the 5′-ends of downstream fragments in reads per million on the EP* reporter. The first nt in the A site codon in the stall motif is designated as zero. b, The results of 3′-RACE showing the 3′-ends of upstream fragments. c, tmRNA tagging sites on the EP* reporter in the wild-type and ΔsmrB strains, corresponding to the residue immediately preceding the tmRNA tag in peptide sequences detected by targeted LC-MS-MS. The relative spectrum count is normalized by the count at the EP* stall site (red) where tmRNA tagging was expected to occur in both the wild-type and ∆smrB strains. The spectrum count corresponds to the mean and the standard deviation of three replicates. The arrow indicates the SmrB cleavage site demonstrated by 5′-RACE. d, 5′-RACE data on the SecM reporter reveal the SmrB cleavage sites as in Fig. 2b, zoomed in to show smaller peaks upstream. e, The distribution of Flag-SmrB in cells treated with 5, 50, or 500 μg/mL erythromycin (ERY) was determined by fractionation over a sucrose gradient and detected with an anti-Flag antibody.
Extended Data Fig. 5 Cryo-EM data processing for the E. coli disome sample.
Shown are the classification scheme, representative micrographs (the scale bar is 500 Å), 2D class averages and the Gold standard Fourier Shell Correlation (GSFSC) curve for the volume containing the 70S stalled ribosome and the 30S of the collided ribosome, as well as the full disome.
Extended Data Fig. 6 Analysis of the E. coli disome structure and comparison of different disome structures.
a, The architecture of the E. coli disome is not compatible with bS1 remaining bound to the stalled ribosome. Aligned models of the 30S subunits of the collided (left) and the stalled (middle) ribosomes are shown in surface representation. The position of bS1 as observed in the collided ribosome is shown in purple and the same position of bS1 in the stalled ribosome is indicated by a dashed line. The clash between bS1 of the stalled ribosome and the 30S subunit of the collided ribosome that would occur upon disome formation is shown on the right. b, Cartoon representation of the individual interactions as they occur at the E. coli disome interface. c, 2D class averages and cryo-EM structure model of an E. coli trisome. d, e, Comparison of the E. coli (E.c.) and B. subtilis (B.s.) disomes displaying full and cut views. Note the smaller space between stalled and collided ribosomes in the B.s. disome interface as illustrated by comparing the positions of uS2 proteins in the zoomed view in c. f, Surface representation of the structural model of the S. cerevisiae disome. g&h, Surface representation of the E. coli and B. subtilis hibernation disomes.
Extended Data Fig. 7 Cryo-EM data processing for the B. subtilis disome and E. coli trisome sample.
a, Shown are the classification scheme, and the Gold standard Fourier Shell Correlation (GSFSC) curves for the final volumes of the B. subtilis disome containing the 70S stalled ribosome and the 70S of the collided ribosome. b, Shown are the 2D class averages, classification scheme, and the Gold standard Fourier Shell Correlation (GSFSC) of the E. coli trisome.
Extended Data Fig. 8 Production of collided and non-collided disomes and relative peak areas of monosomes and disomes in the SmrB nuclease assay.
a, mRNA construct to create the collided E. coli disomes and trisomes and below the sucrose density gradient after in vitro translation. The ribosome stalling site is indicated by an asterisk. b, mRNA construct to create the non-collided disomes that were used in the nuclease assay and below the sucrose density gradient after in vitro translation. c, Relative monosome and disome peak area calculated from the sucrose gradient profiles of the SmrB nuclease assay, showing the mean and standard deviation of three replicates. d, The relative decrease of the area of the disome peak upon addition of SmrB is shown as the mean and standard deviation of three replicates. (The mean difference of the relative disome peak area of collided ribosomes between control and SmrB reaction was set to 1).
Extended Data Fig. 9 Cryo-EM data processing for the E. coli disome sample.
Shown are the classification scheme, representative micrographs (the scale bar is 500 Å), 2D class averages and the Gold standard Fourier Shell Correlation (GSFSC) curve for the respective 3D reconstructions. The segmented density for SmrB is colored according to local resolution.
Extended Data Fig. 10 Structural model of SmrB.
a, Secondary structure of SmrB. The DLH to ALA mutation is indicated. b, AF2 prediction models 1-5 as predicted through the API from the Söding lab. The SMR domain is predicted with high confidence, while the linker to the N-terminal helix appears flexible. c, AF2 prediction of the interaction between SmrB and uS2. For this prediction uS2 was fused to the C-terminus of SmrB with a glycine serine linker (39 copies of GS). The prediction shows the N-terminal helix of SmrB folded back onto uS2. d, Adjustment of the AF2 predicted model of SmrB-uS2. Without adjustment according to the cryo-EM density (as shown in D) the SMR domain would clash with the ribosome. e, Top: Cryo-EM density and adjusted model of the SmrB. Middle: Cryo-EM density and rigid body docked model of the N-terminus of SmrB from the collided 30S onto the stalled 30S. A second copy of SmrB was found anchored to uS2 of the stalled ribosome. However, there was no density for the SMR domain of the second SmrB, indicating a high degree of flexibility due to the lack of another ribosome in front of the stalled one. Bottom: in the control disome without SmrB, there is no density for the N-terminus of SmrB. f, Comparison of the AF2 prediction, the homology model, and the adjusted model of SmrB. Compared to the AF2 prediction, the homology model is missing the two N-terminal helices and most of the loops are slightly different (top). The AF2 prediction almost perfectly matched the cryo-EM density map and the corresponding adjusted model (middle and bottom). Only the catalytic loop (carrying the active site mutations) had to be slightly adjusted to prevent clashes with the mRNA. The N-terminus was adjusted as discussed above. g. During the preparation of this manuscript the AF2 prediction for SmrB (YfcN) became available at the alphafold database at EMBL-EBI. The deposited model resembles our final adjusted model very well including the position of the N-terminus. The confidence of the prediction (pLDDT) is indicated.
Extended Data Fig. 11 Testing the importance of structural interactions for SmrB activity.
a, Examples of operons containing both uS21 and SMR-domain proteins. b, The distribution of Flag-tagged full-length SmrB and a construct with only the SMR domain (residues 88–183) was determined by fractionation over sucrose gradient and detection with an anti-Flag antibody. A non-specific band is marked with *. c, Northern blots using the 3′-probe against the CRP reporters with the short SecM stalling motif in wild-type cells, bL9-deletion strain (∆rplI), and a strain where mCherry is fused to the C-terminus of bL9 (bL9-mCherry). Ethidium bromide staining of 16S rRNA serves as a loading control. d, Northern blots using the 3′-probe against the CRP reporters with the short SecM stalling motif in wild-type cells, a strain where MBP is fused to the N-terminus of uS21, and a strain where GFP is fused to the C-terminus of uS6.
Supplementary information
Supplementary Information
This file contains Supplementary Figs. 1 and 2, which show the uncropped western blots and northern blots corresponding to the main and Extended Data Figures.
Supplementary Table 1
This Excel file contains a list and description of oligonucleotides, plasmids and strains used in this study.
Rights and permissions
About this article
Cite this article
Saito, K., Kratzat, H., Campbell, A. et al. Ribosome collisions induce mRNA cleavage and ribosome rescue in bacteria. Nature 603, 503–508 (2022). https://doi.org/10.1038/s41586-022-04416-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-04416-7
This article is cited by
-
Co-transcriptional gene regulation in eukaryotes and prokaryotes
Nature Reviews Molecular Cell Biology (2024)
-
Riboformer: a deep learning framework for predicting context-dependent translation dynamics
Nature Communications (2024)
-
Learning structural heterogeneity from cryo-electron sub-tomograms with tomoDRGN
Nature Methods (2024)
-
Transient disome complex formation in native polysomes during ongoing protein synthesis captured by cryo-EM
Nature Communications (2024)
-
Tailor made: the art of therapeutic mRNA design
Nature Reviews Drug Discovery (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.