Abstract
Alphaviruses, like many other arthropod-borne viruses, infect vertebrate species and insect vectors separated by hundreds of millions of years of evolutionary history. Entry into evolutionarily divergent host cells can be accomplished by recognition of different cellular receptors in different species, or by binding to receptors that are highly conserved across species. Although multiple alphavirus receptors have been described1,2,3, most are not shared among vertebrate and invertebrate hosts. Here we identify the very low-density lipoprotein receptor (VLDLR) as a receptor for the prototypic alphavirus Semliki forest virus. We show that the E2 and E1 glycoproteins (E2–E1) of Semliki forest virus, eastern equine encephalitis virus and Sindbis virus interact with the ligand-binding domains (LBDs) of VLDLR and apolipoprotein E receptor 2 (ApoER2), two closely related receptors. Ectopic expression of either protein facilitates cellular attachment, and internalization of virus-like particles, a VLDLR LBD–Fc fusion protein or a ligand-binding antagonist block Semliki forest virus E2–E1-mediated infection of human and mouse neurons in culture. The administration of a VLDLR LBD–Fc fusion protein has protective activity against rapidly fatal Semliki forest virus infection in mouse neonates. We further show that invertebrate receptor orthologues from mosquitoes and worms can serve as functional alphavirus receptors. We propose that the ability of some alphaviruses to infect a wide range of hosts is a result of their engagement of evolutionarily conserved lipoprotein receptors and contributes to their pathogenesis.
Similar content being viewed by others
Main
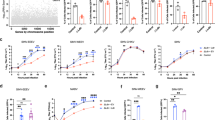
Alphaviruses are enveloped RNA viruses that cause disease in humans ranging from acute febrile illness with rash and arthralgia to lethal encephalitis. Their genomes encode four nonstructural proteins, nsP1–nsP4, and structural proteins, capsid and E3–E2–(6K/TF)–E1). The viral envelope proteins are arranged with icosahedral symmetry and E2–E1 glycoproteins form heterodimers that assemble as 80 trimers that mediate receptor binding and fusion of viral and cellular membranes4,5,6. To use a system that accurately mimics E2–E1 organization, we converted an alphavirus replicon system7 into a DNA-based reporter virus particle (RVP) system in which one plasmid encodes heterologous E3–E2–(6K/TF)–E1 proteins and a second plasmid encodes Ross River virus (RRV) nonstructural proteins, capsid and a reporter (Extended Data Fig. 1a, b). We also generated a library of single guide RNAs (sgRNAs) that target membrane-associated proteins in the human genome (Extended Data Fig. 1c, Supplementary Table 1). We used the library to perform a CRISPR–Cas9 screen for cellular viral receptors using HEK 293T (human kidney epithelial) cells expressing Cas9 (HEK 293T-Cas9) infected with Semliki forest virus (SFV) RVPs. The screen identified VLDLR as the top candidate (Fig. 1a, Supplementary Table 2). VLDLR is a part of the low-density lipoprotein receptor (LDLR) family and mediates endocytosis of lipoproteins and other ligands8.
a, Results of MAGeCK49 analysis for the screen performed with SFV RVPs in HEK 293T-Cas9 cells showing enriched genes on the basis of top robust rank aggregation (RRA) scores. b, Wild-type (WT) cells, VLDLR-knockout (KO) cells and VLDLR-knockout cells transiently transfected with cDNA encoding VLDLR with an N-terminal Flag tag (VLDLR–Flag) were infected with SFV single-cycle RVPs expressing GFP, and infection was measured by fluorescence-activated cell sorting (FACS). VLDLR cell surface expression was monitored by immunostaining (Extended Data Fig. 2b). c, Infection of HEK 293T cells with single-cycle SFV RVPs in the presence of an antibody against VLDLR or a control antibody against human leukocyte antigen (HLA), measured by FACS. d, Infection of Vero cells with SFV or CHIKV single-cycle RVPs expressing GFP in the presence of the indicated antibodies. Cells were imaged by fluorescence microscopy. Scale bar, 100 μm. The experiment was performed twice with representative images shown. e, Infection of Vero cells with replication-competent SINV chimeras expressing GFP and the structural proteins of SFV (SINV-SFV) or CHIKV (SINV-CHIKV) at a multiplicity of infection (MOI) of 1 in the presence of the indicated antibodies. GFP expression was measured by FACS 24 h after infection. f, Infection of HEK 293T cells with GFP-expressing single-cycle RVPs in the presence of receptor-associated protein (RAP) or transferrin (Tf) control, measured by FACS. Data are mean ± s.d. from two experiments (n = 6) (b, c, e, f). One-way ANOVA with Tukey’s multiple comparisons test, ****P < 0.0001 (b, e). Two-way ANOVA with Šídák’s multiple comparison test, ****P < 0.0001 (c, f).
Guide RNAs targeting HSP90B1 and STT3A were also enriched in the screen (Fig. 1a). HSP90B1 encodes an endoplasmic reticulum-resident chaperone that binds the proprotein convertase subtilisin/kexin type 9 serine protease (PCSK9) and prevents PCSK9 from inducing the degradation of LDLR family members9. STT3A encodes the catalytic subunit of the N-oligosaccharyltransferase complex and is also involved in cellular infection by flaviviruses, another group of arthropod-borne viruses that, like alphaviruses, carry positive-sense RNA genomes10,11. STT3A has a role in flavivirus RNA replication and binds to viral nonstructural proteins10,11. Genetic disruption of STT3A in our screen may have, therefore, affected replication through the RRV component of the RVP system and may act downstream of SFV E2–E1-mediated entry.
We focused on exploring the role of human VLDLR as a cellular receptor for SFV. Clonal VLDLR-knockout HEK 293T cells became resistant to infection by GFP-expressing SFV RVPs, and this resistance could be reverted by VLDLR overexpression (Fig. 1b, Extended Data Fig. 2a, b). An antibody against VLDLR, but not a control antibody, blocked infection of HEK 293T cells by SFV RVPs (Fig. 1c). In an experiment using African green monkey kidney (Vero) cells, an anti-VLDLR antibody inhibited the entry of SFV, but not of control RVPs for Chikungunya virus (CHIKV) (Fig. 1d), an alphavirus that uses MXRA8 as a cellular receptor2. The anti-VLDLR antibody also blocked SFV RVP infection of immortalized human cell lines derived from brain, lung, liver, lymphoid, bone and kidney tissues (Extended Data Fig. 2c, d). We generated replication-competent chimeric alphaviruses expressing Sindbis virus (SINV) nonstructural proteins with heterologous structural proteins (capsid and E3–E2–(6K/TF)–E1) and GFP as a reporter. The anti-VLDLR antibody, but not a control antibody, inhibited chimeric SINV–SFV infection of Vero cells (Fig. 1e). Receptor-associated protein (RAP) is a chaperone that binds to some LDLR-related receptors in the endoplasmic reticulum and blocks ligand engagement12. Addition of RAP blocked SFV RVP infection of HEK 293T cells, whereas addition of a control protein did not (Fig. 1f).
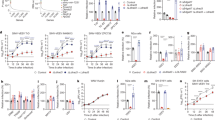
We used on K562 cells13 to determine whether a cell line that is highly resistant to infection could be rendered susceptible by ectopic expression of VLDLR. VLDLR was not detected on the plasma membrane of K562 cells (Extended Data Fig. 2c), which were resistant to SFV RVP infection (Fig. 2a, b). Transduction with VLDLR, however, rendered K562 cells highly susceptible to SFV RVP infection, but not to CHIKV RVP infection (Fig. 2a, b, Extended Data Fig. 3a). Conversely, transduction of K562 cells with MXRA8 rendered them highly susceptible to CHIKV but not to SFV RVP infection (Fig. 2a, b, Extended Data Fig. 3a). Transduction of cells with a control membrane protein had no effect in this assay (Fig. 2b).
a, Infection of wild-type K562 cells or K562 cells expressing indicated proteins infected with SFV or CHIKV single-cycle RVPs expressing GFP. Cells were imaged by fluorescence microscopy. Scale bar, 100 μm. The experiment was performed twice independently with similar results and representative images are shown. b, Infection of wild-type or transduced K562 cells with GFP-expressing SFV or CHIKV single-cycle RVPs measured by FACS. NRP2 is a control membrane protein. c, VLDLR ectodomain and deletion constructs. LBD LA repeats are numbered. d, Infection of K562 cells transduced to express the constructs shown in c or NRP2 with GFP-expressing SFV single-cycle RVPs, measured by FACS. e, Infection of HEK 293T cells with SFV single-cycle RVPs after pre-incubation with VLDLRLBD–Fc or a control NRP2 a1 domain (NRP2a1)–Fc fusion protein, measured by FACS. Cell surface expression of NRP2–Flag, VLDLR–Flag and MXRA8 (a) and VLDLR–Flag variants (d) was confirmed with immunostaining (see Extended Data Fig. 3a). Data are mean ± s.d. from two experiments performed in triplicate (n = 6) (b) or three experiments performed in duplicate (d, e) (n = 6). One-way ANOVA with Tukey’s multiple comparisons test, ****P < 0.0001 (b, d). Two-way ANOVA with Šídák’s multiple comparison test, ****P < 0.0001 (e).
Like other LDLR family members, VLDLR contains an N-terminal ligand-binding domain (LBD) with cysteine-rich repeats (LDLR class A (LA) repeats), a cluster of EGF modules containing a β-propeller domain, and a membrane-proximal O-linked sugar domain8 (Fig. 2c). A construct in which the EGF module containing the β-propeller domain was deleted supported SFV RVP infection, but one in which the LBD was deleted did not (Fig. 2c, d, Extended Data Fig. 3a). Further supporting a role for the LBD as a site of E2–E1 attachment, a VLDLR LBD–Fc fusion protein (VLDLRLBD–Fc), but not an Fc fusion protein comprising a subdomain of an unrelated protein, inhibited SFV RVP infection of HEK 293T cells (Fig. 2e, Extended Data Fig. 4a).
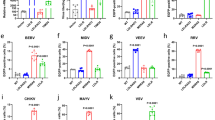
We next sought to determine whether other alphaviruses could bind human VLDLR for cellular entry. Infection by SINV, eastern equine encephalitis virus (EEEV), Venezuelan equine encephalitis (VEEV), western equine encephalitis (WEEV) or CHIKV RVPs was not decreased in VLDLR-knockout HEK 293T cells compared with wild-type cells (Extended Data Fig. 5a). An anti-VLDLR antibody blocked the entry of SFV RVPs into Vero cells but had no effect on the other alphaviruses we tested (Extended Data Fig. 5b). This suggested that some alphaviruses may be able to bind multiple LDLR family members. LDLR, VLDLR and ApoER2 are highly conserved and have superimposable structures, indicating that their corresponding genes evolved from a single ancestor14. ApoER2 and VLDLR have critical roles in brain development and modulate synaptic plasticity in adults14,15,16. ApoER2 is enriched in the brain and can undergo a large number of alternative splicing events that alter the number of LA repeats and ligand-binding properties14,17,18,19,20. We cloned an ApoER2 isoform that contains all seven possible LA repeats (LA1–LA7) (ApoER2iso1) and another that contains only LA1–LA3 (ApoER2iso2), which is thought to be the predominant form14,20 (Extended Data Fig. 6). K562 cells expressed LDLR, but not VLDLR or ApoER2 at the plasma membrane (Extended Data Figs. 2c, 3b). SFV, EEEV and SINV RVPs infected K562 cells transduced with VLDLR or ApoER2 isoforms to varying degrees (Extended Data Fig. 5c, d). Addition of RAP or a soluble VLDLR LBD (sVLDLRLBD) protein that does not contain an Fc segment (which we avoided because K562 cells express Fc receptors) blocked E2–E1-mediated infection of K562 cells expressing VLDLR or ApoER2iso2, confirming that infection was a result of ectopic lipoprotein receptor expression (Extended Data Fig. 5e). VLDLR or ApoER2 expression did not affect CHIKV, VEEV or WEEV RVP infection of K562 cells (Extended Data Fig. 5f). sVLDLRLBD blockade of SFV, EEEV and SINV RVP infection of K562 cells overexpressing ApoER2iso2 (Extended Data Fig. 5e) suggest that VLDLR and ApoER2 bind an overlapping site on the E2–E1 proteins of these alphaviruses.
To confirm the interaction of alphavirus E2–E1 proteins with the VLDLR LBD, we transfected HEK 293T cells with plasmids encoding different alphavirus E3–E2–(6K/TF)–E1) proteins and conducted cell surface staining experiments with Fc fusion proteins (Fig. 3a, Extended Data Fig. 4a). VLDLRLBD–Fc-bound cells transfected with the E3–E2–(6K/TF)–E1 proteins of SFV, EEEV and SINV, but not cells transfected with CHIKV E3–E2–(6K/TF)–E1 (Fig. 3a). Conversely, the MXRA8 ectodomain (MXRA8ect)–Fc fusion protein, but not VLDLRLBD–Fc, bound cells transfected with CHIKV E3–E2–(6K/TF)–E1 (Fig. 3a).
a, Cell surface expression of VLDLRLBD–Fc or MXRA8ect–Fc in HEK 293T cells transfected with plasmids encoding alphavirus E3–E2–(6K/TF)–E1 proteins. PE, R-phycoerythrin. b, BLI-based binding analysis of VLPs to sensor tips coated with VLDLRLBD–Fc or MXRA8ect–Fc after pre-dipping into buffer or solution containing RAP or transferrin (Tf). The maximal response value is plotted. Sensorgrams are shown in Extended Data Fig. 4c. c, xy slice and 3D volume renderings of representative images of WGA (green)-stained transduced K562 cells incubated with fluorescently labelled VLPs (pink) imaged by live-cell confocal microscopy after co-incubation of cells and VLPs at the indicated temperatures (see Extended Data Fig. 6). Scale bars, 5 μm. d, Number of VLPs bound to individual cell membranes (membr.) or found in the cytoplasm (cyto.) of individual cells at the indicated temperatures (see Extended Data Fig. 8). Data are mean ± s.d. from two experiments performed in triplicate (n = 6); two-way ANOVA with Šídák’s multiple comparison test, ****P < 0.0001 (a). Mean of values obtained from two experiments; one-way ANOVA with Tukey’s multiple comparisons test, ****P < 0.0001; ***P = 0.0003 (d).
To determine whether alphavirus E2–E1 proteins directly bind the LBDs of VLDLR and ApoER2 without a requirement for associated lipoproteins, we generated and purified virus-like particles (VLPs), which replicate the structure of native virions21,22 (Extended Data Fig. 1d). When we examined VLPs by negative-stain electron microscopy, we observed no associated lipoproteins (Extended Data Fig. 4b). Mass spectrometry did not reveal substantial amounts of lipoprotein-associated peptides in purified VLP samples as compared to a VLDL control (Supplementary Table 3). In biolayer interferometry (BLI)-based experiments, VLDLRLBD–Fc, but not MXRA8ect–Fc, captured SFV, SINV and EEEV VLPs (Fig. 3b, Extended Data Fig. 4c). We also generated an ApoER2iso1 LBD–Fc fusion protein (ApoER2LBDiso1–Fc) (Extended Data Fig. 4a). ApoER2LBDiso1–Fc captured SFV, EEEV and SINV VLPs (Fig. 3b, Extended Data Fig. 4c). Addition of RAP, but not a control protein, specifically blocked VLP binding to VLDLRLBD–Fc and ApoER2LBDiso1–Fc (Fig. 3b, Extended Data Fig. 4c). Thus the LBDs of VLDLR and ApoER2 interact directly with alphavirus E2–E1 proteins.
We next turned to confocal microscopy to determine whether the expression of VLDLR or ApoER2iso2 (chosen because this shorter form is predominant14,20) enables cell surface binding and internalization of fluorescently labelled VLPs. We incubated labelled VLPs with transduced K562 cells that had also been treated with heparinase and stained with wheat germ agglutinin (WGA) to visualize cell membranes (Fig. 3c, Extended Data Figs. 7, 8). Expression of VLDLR, but not MXRA8, promoted the binding of labelled SFV VLPs to cell surface membranes, and more particles were detected in the cytoplasm of cells at 37 °C than at 4 °C, suggesting internalization (Fig. 3c, d). We also observed an increased number of SFV VLPs in the cytoplasm of cells expressing ApoER2iso2 at 37 °C. The expression of VLDLR and ApoER2iso2 promoted cell surface binding of EEEV VLPs, and more particles were again detected in the cytoplasm of cells expressing VLDLR or ApoER2iso2 at 37 °C (Fig. 3d). We detected cell surface binding and internalization of SINV VLPs on cells expressing VLDLR and ApoER2, but the magnitude of the effects was more modest than those observed with SFV and EEEV VLPs (Fig. 3d).
SFV is neuropathogenic in young laboratory mice23 and has also caused fatal encephalitis in an exposed laboratory worker, although this individual had chronic purulent bronchitis and may have been immunocompromised24. We sought to determine whether VLDLRLBD–Fc or RAP could prevent SFV RVP infection of mouse primary cortical neurons and of human neurons differentiated from induced pluripotent stem (iPS) cells25. VLDLRLBD–Fc and RAP, but not a control protein, blocked SFV RVP infection of mouse cortical neurons and human iPS cell-derived neurons (Fig. 4a, b, Extended Data Fig. 9a–d).
a, Infection of mouse primary cortical neurons with GFP-expressing SFV single-cycle RVPs in the presence of the indicated proteins. Cells were imaged by fluorescence microscopy. Scale bars, 100 µm. Phase-contrast images are shown in Extended Data Fig. 9c. The experiment was performed twice, and representative images are shown. b, Quantification of infection in the experiment shown in a using a live cell imaging system (see Methods). c, d, Ten-day-old mice were administered a VLDLRLBD–Fc fusion protein or an isotype control antibody intraperitoneally 6 h before intraperitoneal inoculation with 100 PFU (c) or 1,000 PFU (d) of SFV A774. Survival of the mice was monitored daily. e, Infection of wild-type K562 or K562 cells transduced with VLDLR orthologues with single-cycle RVPs expressing GFP. f, Infection of wild-type K562 cells or K562 cells transduced with ApoER2 orthologues with single-cycle RVPs expressing GFP. Cell surface expression of constructs used in e and f, was confirmed by immunostaining (Extended Data Fig. 3). Data are mean ± s.d. from two experiments performed in triplicate (n = 6), except for the RAP experiment in b, which was performed once in triplicate and once in duplicate (n = 5). One-way ANOVA with Tukey’s multiple comparisons test, ****P < 0.0001 (b, e, f). Survival data (c, d) are from two independent experiments; in c: PBS n = 10, VLDLRLBD–Fc n = 10 isotype control n = 9 mice; in d: PBS, n = 11, VLDLRLBD–Fc n = 12, isotype control n = 11 mice. log-rank (Mantel–Cox) test comparing VLDLRLBD–Fc or isotype control to PBS, ****P < 0.0001 (c, d); or isotype control to PBS, P = 0.4745 (c) or P > 0.9999 (d); NS, not significant. H. sapiens, Homo sapiens.
We next tested wild-type, replication-competent strains of SFV (A774), EEEV (FL-939-39, and SINV (Ar Mg812) in a multi-step viral replication assay using transduced K562 cells. Ectopic expression of VLDLR and ApoER2 isoforms resulted in faster kinetics and increased levels of viral replication, and we observed a three-log increase in viral replication for SFV and an almost five-log increase with EEEV (Extended Data Fig. 9e). The effect was less pronounced but nonetheless significant with SINV infection, consistent with the moderate phenotype we observed with SINV RVPs in infectivity assays with K562 cells expressing VLDLR and ApoER2 (Extended Data Fig. 5c, d) or VLP cell binding and internalization assays (Fig. 3c, d).
When infected with SFV strain A774, neonatal mice, but not adult mice, rapidly die from fulminant encephalitis26,27,28,29,30. Because VLDLR and ApoER2 are important for the development of the central nervous system, mice that are deficient in both receptors have ataxia, severe cognitive deficits and early lethality31, thus limiting our ability to carry out in vivo studies in double-knockout mice. We instead used VLDLRLBD–Fc as a blocking agent for in vivo studies. This protein should block VLDLR- and ApoER2-dependent cellular entry, as sVLDLRLBD blocking experiments suggest that alphavirus E2–E1 proteins use the same surface to bind to both receptors (Extended Data Fig. 5e). VLDLRLBD–Fc neutralized SFV A774 in a plaque assay (Extended Data Fig. 5g). When ten-day-old mice were inoculated with SFV A774 six hours after receiving phosphate-buffered saline or an isotype control IgG, all mice rapidly succumbed to infection within three days of viral challenge. However, 100% of mice treated with VLDLRLBD–Fc 6 h before viral challenge were still alive 3 days after challenge (Fig. 4c, d). Although all VLDLRLBD–Fc-treated mice eventually succumbed to infection (became moribund, meeting euthanasia criteria) on day 8 (100 plaque-forming units (PFU) challenge dose group) or day 7 (1,000 PFU challenge dose group), the isotype control-treated mice all became moribund significantly faster, by day three. Because VLDLRLBD–Fc could have, in principle, been cleared from circulation by associating with lipoproteins (in addition to binding virus), further studies will be required to determine whether repeated administration would provide additional therapeutic benefit in this model system for otherwise rapidly fatal viral encephalitis.
The LBDs of VLDLR and ApoER2 are mostly conserved even among highly divergent species (Extended Data Figs. 6, 10). We transduced K562 cells with various VLDLR and ApoER2 orthologues and—taking advantage of the ability of RAP to interact with highly divergent lipoprotein receptor orthologues32,33 but not with LDLR when added exogenously34—we used RAP to monitor orthologue cell surface expression (Extended Data Fig. 3c). SFV RVPs infected K562 cells that expressed equine (Equus caballus) and avian (Sturnus vulgaris) VLDLR orthologues (Fig. 4e). SFV and EEEV infected cells that expressed mosquito (Aedes aegypti and Aedes albopictus) VLDLR orthologues (lipophorin receptor 1) (Fig. 4e). Remarkably, SFV RVPs could also infect cells overexpressing a Caenorhabditis elegans VLDLR orthologue, a receptor with a role in regulating the worm’s intestinal lipid content35,36 (Fig. 4e). Murine, equine and avian ApoER2 orthologues supported entry of SFV, EEEV and SINV RVPs to varying degrees (Fig. 4f, Extended Data Fig. 10c).
ApoER2 is almost exclusively expressed in the central nervous system14. Accordingly, we did not detect ApoER2 on the surface of HEK 293T, Vero or K562 cells (Extended Data Fig. 3b). These findings may explain why incubation with an antibody against VLDLR is sufficient to block SFV E2–E1-mediated infection of multiple cell lines (Fig. 1c–e, Extended Data Fig. 2d). Whereas SFV primarily depends on VLDLR for entry into several cell types, EEEV and SINV—although they can bind VLDLR and ApoER2—can enter HEK 293T and Vero cells through independent pathways, as genetic disruption of VLDLR in HEK 293T cells, or treatment of Vero cells with an antibody against VLDLR, did not decrease EEEV or SINV RVP infection of these cells (Extended Data Fig. 5a, b). NRAMP2 is a possible alternative receptor for SINV1. Other LDLR-family members that we did not test, including LRP1, LRP1b, LRP2 and LRP4, could also have roles in alphavirus entry. An anti-VLDLR antibody had a modest effect on SFV entry into U2OS cells, a human bone-derived cell line (Extended Data Fig. 2d), also suggesting the presence of alternative SFV receptors on this cell type. SFV is reported to partially depend on MXRA8 for entry into mouse cells2. Human MXRA8, however, is an unlikely alternative receptor for SFV on U2OS cells, as we did not detect SFV RVP entry into K562 cells overexpressing human MXRA8 (Fig. 2a, b, Extended Data Fig. 5c, d), nor did we detect SFV VLP binding to these cells (Fig. 3d) or to human MXRA8ect–Fc in BLI experiments (Fig. 3b).
SFV causes encephalitis in horses, mice, rats, rabbits and guinea pigs, and SINV also causes age-dependent encephalitis in mice26,37,38. Because SFV, SINV and EEEV can cause encephalitis in humans or animals, binding to VLDLR or ApoER2 could contribute to their neuropathogenesis. VEEV has recently been shown to bind low-density lipoprotein receptor class A domain-containing 3 (LDLRAD3) as a receptor3. While LDLRAD3A is found in vertebrates, it does not have an apparent orthologue in mosquitos3. However, every VLDLR orthologue we tested supported SFV E2–E1-mediated infection, including that of C. elegans, which is separated by 109 years of evolutionary divergence from humans (Fig. 4e, Extended Data Fig. 10c). Furthermore, only the entry of VEEV RVPs, but not that of SFV, EEEV or SINV RVPs, was enhanced by overexpression of LDLRAD3 on K562 cells, suggesting that the VEEV E2–E1 proteins evolved the ability to only recognize this specific receptor that is structurally homologous to LDLR family members (Extended Data Fig. 5h). Our data, therefore, reveal that similarities in alphavirus E2–E1 protein structure correlate with structural homology in receptors from organisms as evolutionarily distant from humans as worms.
The lipoprotein receptor gene family appeared in an evolutionary burst at the advent of multicellular life and has maintained a remarkable degree of conservation throughout evolutionary history36. The ability of some alphaviruses to bind LDLR family members probably accounts for their extensive tissue and species tropism. Many viruses, including vesicular stomatitis virus34, minor group rhinoviruses39,40, subgroup A Rous sarcoma virus41, several Flaviviridae family members42,43,44, VEEV3 and Rift Valley fever virus45, have been reported to bind LDLR-repeat-containing receptors to enter cells. LDLR is also a receptor for Clostridium difficile toxin A46. LDLR family members may, therefore, represent evolutionary conserved ‘hotspots’ for interfacing with pathogens. Pathogens, in turn, may be driving sequence divergence of the receptor LBDs. A similar evolutionary arms race has been described with the iron-uptake protein transferrin receptor 1, which is also recurrently targeted by pathogens for cellular entry47,48.
Our studies help answer, in part, the longstanding question of how some alphaviruses can infect a wide range of organisms. They further suggest that strategies targeting multiple cellular receptors, and possibly multiple receptor binding sites on virions, may be required to effectively limit the cellular entry of some pathogenic alphaviruses.
Methods
Cells and viruses
We maintained HEK 293T (human kidney epithelial, ATCC CRL-11268), 293FT (Thermo Fisher Scientific R70007), Vero (Cercopithecus aethiops kidney, ATCC CCL-81), U2OS (human bone, ATCC HTB-96), A549 (human lung epithelial, ATCC CCL-185), SVG-A (human astroglial, provided by T. Kirchhausen), and Huh7 cells (provided by F. Zhang) in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 10% (v/v) fetal bovine serum (FBS), 25 mM HEPES (Thermo Fisher Scientific), and 1% (v/v) penicillin-streptomycin (Thermo Fisher Scientific). We maintained Jurkat clone E6-1 (human lymphoblast, ATCC TIB-152) and K562 (human chronic myelogenous leukemia, ATCC CCL-243) cells in RPMI (Thermo Fisher Scientific) supplemented with 10% (v/v) FBS, 25 mM HEPES, and 1% (v/v) penicillin-streptomycin. We maintained SK-N-SH (human brain, ATCC HTB-11) and EBC-1 (human squamous cell lung carcinoma, provided by T. Kirchhausen) in Eagle’s minimum essential medium (EMEM, Sigma) supplemented with 10% (v/v) FBS, 25 mM HEPES, and 1% (v/v) penicillin-streptomycin. We maintained Expi293F cells (Thermo Fisher Scientific A14527) in Expi293 Expression Medium (Thermo Fisher Scientific) supplemented with 1% (v/v) penicillin-streptomycin. We maintained BHK-21 cells (ATCC CCL-10) in DMEM supplemented with 10% (v/v) fetal bovine serum and 1% (v/v) penicillin-streptomycin. Cell lines were not authenticated. We confirmed the absence of mycoplasma in all cell lines through monthly testing using an e-Myco PCR detection kit (Bulldog Bio 25234).
The following wild-type replication-competent alphaviruses were used: SINV (strain DAK Ar Mg812) and EEEV (strain FL-93-939). These viruses were propagated in Vero CCL-81 cells and titrated by standard plaque assays. For work with unmodified replication-competent SFV, we rescued a molecular clone using a plasmid encoding SFV A774 (pCMV-A774wt)50 obtained from A. Merits. The SFV clone was rescued by electroporating 10 μg of pCMV-A774wt plasmid into BHK-21 cells (220 V, 975 μF, one pulse in a cuvette with a 4-mm electrode gap) using a BTX-Harvard Apparatus ECM 830 Square Wave Electroporator (Harvard Apparatus). After incubation for 24 h at 37 °C, the stock of rescued virus (P0 stock) was collected and titrated in a plaque assay on Vero cells. To obtain P1 stock, confluent BHK-21 cells grown on T75 flasks were infected with P0 stock at an MOI of 5. At 24 h post infection, the supernatant (P1 stock) was collected, and virus titre was determined on Vero cells.
Primary mouse cortical neuron culture and infection assays
Mouse experiments were approved at Harvard Medical School under the Harvard Medical School Institutional Animal Care and Use Committee protocol number IS00000054. Mouse (C57BL/6J) primary cortical neurons were dissociated and cultured using the Papain Dissociation System (Worthington Biochemical LK003153) as previously described51. In brief, postnatal day 0 (P0) C57BL/6J mice were euthanized and mouse cortices were collected in cold Earle’s balanced salt solution (EBSS) and resuspended in 2.5 ml of warmed EBSS supplemented with papain (20 units ml−1) and DNase (2000 units ml−1). Following a 12 min incubation at 37 °C, cortices were triturated using fire polished glass Pasteur pipettes. Samples were centrifuged (2,000g for 5 min) to pellet cells, and then resuspended in 1.6 ml of suspension media (1.375 ml EBSS, 150 µl albumin-ovomucoid inhibitor (10 mg ml−1 in EBSS), and 75 µl DNase (2,000 units ml−1). This solution was then layered on top of a 2.5 ml solution of albumin-ovomucoid inhibitor (10 mg ml−1 in EBSS) to create a continuous density gradient and the samples were centrifuged at 1,000 rpm for 5 min. The supernatant (gradient) was discarded, and pelleted neurons were collected in pre-warmed Neurobasal Plus medium (Thermo Fisher Scientific) supplemented with 200 mM l-glutamine (Thermo Fisher Scientific) and 1× B-27 (Thermo Fisher Scientific) with 1% (v/v) penicillin-streptomycin (Thermo Fisher Scientific). Cells were plated in 24 well glass bottom dishes (Cellvis 24 well plate 1.5 glass bottom cover dish) (P241.5HN) for imaging experiments at a density of 4 × 105 cells per well. After neurons had been cultured for 7 d, we pre-incubated SFV RVPs with VLDLRLBD–Fc fusion proteinsor RAP in culture media containing 5 µg ml−1 polybrene for 30 min at 37 °C. We then added SFV RVP and Fc fusion proteins or SFV RVP and RAP mixtures to cells. Cells were imaged every 2 h for 24 h using the Incucyte S3 Live Cell Imaging system (Sartorius) with Incucyte S3 Software v2018B (Sartorius) using the following objectives: 20×/0.45 Plan Fluor (4465), 10×/0.3 Plan Fluor (4464). GFP-positive neurons were scored as cells with a threshold signal greater than 5 green calibrated units (GCU) above background, using a Top-hat background subtraction method. To calculate the percent positive cells, at the time point of 22 h post-infection, the area of GFP signal above background was divided by the total area covered by cells under phase contrast and was multiplied by 100. We calculated relative infection as follows: Relative infection (%) = (% GFP-positive cells in the presence of antibody or Fc fusion protein or RAP)/(% GFP-positive cells in the absence of antibody or Fc fusion or RAP) × 100%.
Induced pluripotent stem cell lines
The generation of cell lines from human iPS cells was approved by the institutional review board (IRB) of Brigham & Women’s Hospital (IRB protocol 2015P001676). iPSCs were generated from peripheral blood mononuclear cells (PBMCs) from the Religious Order Study (ROS) and Memory and Aging Project (MAP) cohort using the Sendai virus reprogramming method as previously described25. iPS cells underwent a rigorous quality check procedure that includes a sterility check, mycoplasma testing, karyotyping and pluripotency assays performed by the New York Stem Cell Foundation (NYSCF). iPS cells were maintained using StemFlex Medium (Thermo Fisher Scientific). For this study, two cell lines (one male and one female) were used for induced neuron differentiation.
Induced neuron differentiation
Induced neurons were generated as previously described52 with minor modifications that have also been previously described25. In brief, iPS cells were plated at a density of 95,000 cells per cm2 on a growth factor reduced Matrigel basement membrane matrix (Corning) coated plate, then were transduced with three lentiviruses: pTet-O-NGN2-puro (Addgene plasmid #52047, a gift from M. Wernig)52, Tet-O-FUW-EGFP (Addgene plasmid 30130, a gift from M. Wernig)53, and FUdeltaGW-rtTA (Addgene plasmid 19780, a gift from K. Hochedlinger)54. The cells were then dissociated with StemPro Accutase Cell Dissociation Reagent (Thermo Fisher Scientific) plated at 200,000 cells per cm2 using StemFlex and ROCK inhibitor Y-27632 (Stemcell Technologies) (10 µM) (day 0). From day 1 to day 3, the media was gradually switched from KSR media (KnockOut DMEM (Thermo Fisher Scientific), 15% (v/v) KnockOut Serum Replacement (Thermo Fisher Scientific), 1× MEM non-essential amino acids solution (Thermo Fisher Scientific), 55 µM β-mercaptoethanol (Thermo Fisher Scientific), 1× GlutaMAX (Life Technologies)) to N2B media (DMEM: Nutrient Mixture F-12 (DMEM/F12) (Thermo Fisher Scientific), 1× GlutaMAX (Life Technologies), 1× N2 Supplement-B (Stemcell Technologies), 0.3% (v/v) dextrose (d-(+)-glucose) (Sigma)). Day 1 media contained 100% (v/v) KSR, day 2 media contained 50% (v/v) KSR and 50% (v/v) N2B, and day 3 media contained 100% (v/v) N2B. Doxycycline (2 µg ml−1) (Sigma) was added from day 1 to the end of the differentiation, and puromycin (5 µg ml−1) (Gibco) was added from day 2 to the end of the differentiation. On day 3, B27 supplement (1:100) (Life Technologies) was added. On day 4, cells were replated at 50,000 cells per cm2 and from day 4 to the end of differentiation (day 21), cells were cultured with NBM media (Neurobasal medium (Thermo Fisher Scientific), 0.5× MEM-NEAA (Thermo Fisher Scientific), 1× GlutaMAX 0.3% dextrose (d-(+)-glucose) (Sigma) supplemented with 1:50 B27 + BDNF, GDNF, CNTF (10 ng ml−1) (PeproTech)) with media replaced every 2 to 3 days. After neurons had been cultured for an additional 14 d, we pre-incubated SFV RVPs with VLDLRLBD–Fc fusion proteins or RAP in culture media containing 5 µg ml−1 polybrene for 30 min at 37 °C. We then added SFV RVP and Fc fusion proteins or SFV RVP and RAP mixtures to cells. Cells were imaged every 2 h for 24 h using the Incucyte S3 Live Cell Imaging system and relative infection based on GFP expression at the time point of 22 h post-infection was calculated as described above in experiments performed with primary mouse cortical neuron cultures.
Reporter virus particle generation
To create a two-component RVP system, we used elements from an RRV replicon plasmid (pRR64)7 provided by R. Kuhn (Purdue University). We removed the SP6 promoter and replaced it with a CMV/T7 promoter cassette. We also removed RRV E3–E2–(6K/TF)–E1 sequence from pRR64 and replaced it with reporter gene (CD20) or turbo GFP downstream of the capsid gene sequence and preceded by a 2A ‘self-cleaving’ peptide derived from porcine teschovirus-1, and included a bGH termination signal after the 3'UTR poly-A tail. RVPs were generated by providing in trans the modified pRR64 plasmid and a pCAGGS vector expressing the heterologous alphavirus E3–E2–(6K/TF)–E1 proteins with a start codon upstream of E3. pCAGGS E3–E2–(6K/TF)–E1 expressor plasmids for CHIKV (strain 37997, GenBank AY726732.1) and EEEV (Florida 91-469, Q4QXJ7.1) have been previously described55. pCAGGS E3–E2–(6K/TF)–E1 expressor plasmids for SFV (SFV4, AKC01668.1), SINV (Toto1101 T6P144, AKZ17594.1), WEEV (71V-1658, NP_640331.1), and VEEV (TC-83, AAB02517.1) were subcloned for this study. We used Lipofectamine 3000 (Invitrogen) to transfect 293FT cells using the manufacturer’s protocol and replaced media with Opti-MEM (Thermo Fisher Scientific) supplemented with 5% (v/v) FBS, 25 mM HEPES, and 5 mM sodium butyrate 1 d post-transfection. We collected supernatants 3 d post-transfection. Supernatants were then centrifuged at 1,000 rpm for 5 min, filtered using a 0.45 µm filter, and frozen at −80 °C for storage.
To purify RVPs for SDS–PAGE analysis, we collected supernatants 24 and 48 h (EEEV and SINV) or 48 h (SFV) post-transfection. We clarified supernatants by centrifugation at 3,000g for 10 min. We then performed polyethylene glycol (PEG)-precipitation by mixing clarified supernatants to a final concentration of 7% (v/v) PEG 6000 and 2.3% (v/v) NaCl and incubated samples at 4 °C for 4 h. Precipitates were pelleted by centrifugation at 4,000g for 30 min and resuspended in phosphate-buffered saline (PBS). We loaded resuspended RVPs onto a 20–70% continuous sucrose gradient and centrifuged samples at 210,000g for 1.5 h. We collected VLP bands and then used 100-kDa Amicon filters (Sigma UFC910096) to buffer exchange samples into PBS and to concentrate samples. Samples were analysed by SDS–PAGE using a 4–15% Mini-PROTEAN TGX Stain-Free Protein Gel (Bio-Rad 4568083), with proteins visualized using a stain-free gel imaging system (Bio-Rad ChemiDoc). Uncropped, unprocessed images of scanned gels are provided in Supplementary Fig. 1.
sgRNA library design, screening, and data analysis
We generated a list of membrane or membrane-associated proteins by examining list of proteins determined to be on the cell surface by mass spectrometry56 or predicted bioinformatically to be cell surface associated57,58. We also obtained a list of genes that encode proteins predicted to be on endosomes, lysosomes, vesicles or the cell surface by UniProt (https://www.uniprot.org). To compile a comprehensive list of genes using these resources, we included genes encoding proteins that were identified to be cell surface associated by mass spectrometry and then added any other gene that was predicted to be on cell surface by at least one of the other resources. The final list of genes is included in Supplementary Table 1. A CRISPR knockout library containing 10 sgRNAs per gene was generated by Desktop Genetics. We amplified the library in Endura ElectroCompetent cells (Lucigen 60242) as previously described59. We packaged the sgRNA plasmid library in HEK 293T cells through co-transfection of the lentiGuide-Puro vector (provided by F. Zhang, Addgene #52963)60, psPAX2 (provided by D. Trono, Addgene #12260) and pMD2.G (provided by D. Trono, Addgene #12259) using Lipofectamine 3000 (Invitrogen) and following the manufacturer’s instructions. Supernatants were collected 1 and 2 d post-transfection, pooled, clarified by centrifugation (1,200 rpm for 5 min), filtered through a 0.45 µm membrane, and stored at −80 °C.
To generate a clonal HEK 293T cell line that expresses Streptococcus pyogenes Cas9 (HEK 293T-Cas9), we transduced cells with lentiCas9-blast (provided by F. Zhang, Addgene #52962)60, selected transduced cells with blasticidin, and isolated clones by limiting dilution. We selected a HEK 293T-Cas9 clone with high Cas9 activity after clonal dilution by transfecting cells with pXPR_011 vector (provided by J. Doench and D. Root, Addgene #59702)61, which expresses GFP and an sgRNA against GFP, and monitoring for low GFP expression by FACS. We selected a clone that had roughly an 80% decrease in GFP signal as compared to WT HEK 293T cells. We expanded the HEK 293T-Cas9 cells and transduced cells (150 × 107) with the CRISPR sgRNA lentivirus library at a multiplicity of infection (MOI) of 0.3. We began selection of sgRNA containing cells with puromycin at 1 µg ml−1 1 d post-transduction. Seven to 10 d post-selection, we infected cells with SFV RVPs expressing CD20, aiming for 80–90% infected cells as monitored by an anti-CD20 APC conjugate antibody (Miltenyi Biotec Clone LT20 130-113-370) used at 1:50 dilution using separate HEK 293T-Cas9 cells that had not been exposed to sgRNAs. Three days post RVP infection, we depleted infected cells using anti-CD20 MicroBeads (Miltenyi Biotec 130-091-104) and following the manufacturer’s protocol. We chose to use anti-CD20 MicroBeads to deplete infected cells, instead of flow cytometry to separate cells based on GFP expression, because the former isolation strategy could be performed in a biosafety cabinet rather than requiring several hours of instrument time on a flow cytometer. To improve the signal-to-noise ratio, we expanded non-infected cells and repeated infection with SFV RVPs expressing CD20 for an additional two rounds. We extracted genomic DNA from uninfected cells and control HEK 293T-Cas9 cells that had been exposed to sgRNAs and puromycin for the duration of the experiment but had not been infected with RVPs. We amplified sgRNA sequences and determined sgRNA content using next generation sequencing on an Illumina MiSeq. We determined sgRNA sequences targeting specific genes after removing tag sequences. We analysed sequences for gene enrichment using MAGeCK49.
Genetic knockout validation
For disrupting genes using CRISPR–Cas9, we used paired sgRNAs62 with a nuclease to introduce ~200-bp deletions in target genes. We used Lipofectamine 3000 (Invitrogen) and followed the manufacturer’s protocol to co-transfect HEK 293T cells with the lentiGuide-Puro vector (Addgene #52963)60 expressing sgRNAs and the lentiCas9-blast plasmid (Addgene #52962)60. Two days post-transfection, we split cells and started selection with blasticidin at 10 µg ml−1 and puromycin at 1 µg ml−1. We returned cells to normal media for replication 3 d post-transfection and isolated individual clones using clonal dilution. We isolated genomic DNA from clonal cells and used a genotyping PCR to confirm successful deletion and confirmed lack of cell surface VLDLR expression using the mouse anti-human VLDLR antibody 1H10 (GeneTex GTX79552).
sgRNA sequences for VLDLR disruption were as follows: VLDLR sgRNA-1: 5′-CACCGCGACCAATCTGATGAGTCCC-3′; VLDLR sgRNA-2: 5′-AAACGGGACTCATCAGATTGGTCGC-3′; VLDLR sgRNA-3: 5′-CACCGACTGGAGCAGGTGAACTCGT-3′; VLDLR sgRNA-4: 5′-AAACACGAGTTCACCTGCTCCAGTC-3′.
Genotyping primer sequences were as follows: primer 1: 5′-CCATTGTAGCCTTTAAGTTGGG-3′; primer 2: 5′-TCCTCACATTCAAATTGGTCAG-3′.
SINV chimera generation
A plasmid encoding infectious SINV expressing GFP (pTE3′2J)63 was provided by R. Andino. To generate plasmid launched SINV chimeric recombinant alphaviruses, we replaced the Sp6 promoter located at the 5′ end of the SINV insert with a CMV/T7 promoter cassette and introduced at the 3′ end the hepatitis delta virus (HDV) ribozyme and SV40 poly(A) sequences. To generate replication-competent SINV chimeric viruses, we then replaced the genes encoding SINV structural proteins (capsid and E3–E2–(6K/TF)–E1) with the structural proteins of CHIKV (strain 37997, GenBank AY726732.1) or SFV (SFV4, AKC01668.1). We transfected HEK 293T cells using Lipofectamine 3000 (Invitrogen) following the manufacturer’s protocol. We replaced media 12 h post-transfection with Opti-MEM (Thermo Fisher Scientific) with 5% (v/v) FBS and 25 mM HEPES. We collected the supernatant 48 h after transfection, filtered these through a 0.45 µm filters, and froze supernatants at −80 °C for storage. Chimeras were titred on Vero (CCL-81) cells by plaque assay.
Expression and purification of virus-like particles
To produce SINV and SFV VLPs, we subcloned the structural proteins (capsid and E3–E2–(6K/TF)–E1) of SINV (Strain T6P144, GenBank AKZ17594.1) and SFV (SFV4, AKC01668.1) into the mammalian expression vector pHLSec64. We used previously described vectors to produce CHIKV21 and EEEV22 VLPs. We transfected HEK 293T cells with Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. We purified VLPs as previously described65. In brief, we collected supernatant 24 h and 48 h post-transfection and clarified these by centrifugation at 3,000g for 10 min. We then performed PEG precipitation by mixing clarified supernatants to a final concentration of 7% (v/v) PEG 6000 and 2.3% (v/v) NaCl and incubated at 4 °C overnight. Precipitates were pelleted by centrifugation at 4,000g for 30 min and resuspended in PBS. We loaded resuspended VLPs onto a 20–70% continuous sucrose gradient and centrifuged samples at 210,000g for 1.5 h. We collected VLP bands and buffer exchanged using a 100-kDa Amicon filter (Sigma). VLPs were stored at 4 °C in Tris buffered saline (TBS), pH 7.4, containing 2 mM CaCl2 (for BLI experiments) or in PBS (for electron microscopy experiments) and not frozen. We confirmed particle integrity and the absence of degradation products using SDS–PAGE (Extended Data Fig. 1d) in addition to negative-stain electron microscopy (Extended Data Fig. 4b). VLPs were always used within seven days of purification.
Protein sequence analysis by LC–MS/MS
Protein sequence analysis by LC–MS/MS of purified alphavirus VLPs and human VLDL (Sigma LP1) was performed at the Taplin Biological Mass Spectrometry Facility at Harvard Medical School. Prior to analysis, samples were digested in trypsin followed by a reverse phase clean up. Samples were then dried in a Vacufuge (Eppendorf) concentrator for ~1 h and stored at 4 °C until analysis. On the day of analysis, samples were reconstituted in 10 µl of high-performance liquid chromatography (HPLC) solvent A (2.5% (v/v) acetonitrile, 0.1% (v/v) formic acid). A nano-scale reverse-phase HPLC capillary column was created by packing Accucore 2.6 µm C18 spherical silica beads (Thermo Fisher Scientific) into a fused silica capillary (100 µm inner diameter × ~30 cm length) (Polymicro Technologies) with a flame-drawn tip66. After equilibrating the column each sample was loaded using a FAMOS autosampler (LC Packings) onto the column. A gradient was formed, and peptides were eluted with increasing concentrations of solvent B (97.5% (v/v) acetonitrile, 0.1% (v/v) formic acid). As peptides eluted, they were subjected to electrospray ionization and then entered into an LTQ Orbitrap Velos Pro ion-trap mass spectrometer (Thermo Fisher Scientific). Peptides were detected, isolated, and fragmented to produce a tandem mass spectrum of specific fragment ions for each peptide. Peptide sequences (and hence, protein identity) were determined by matching protein databases with the acquired fragmentation pattern using the software program SEQUEST version 28 rev 13 (Thermo Fisher Scientific)67. All databases include a reversed version of all the sequences and the data was filtered to between a one and two percent peptide false discovery rate.
Labelling of virus-like particles
We purified VLPs as described above with the exception that we first buffer exchanged particles into 0.1 M Sodium Bicarbonate (pH 8.3) and diluted them to a concentration of 1 mg ml−1 for labeling. Immediately before use, Alexa Fluor 647 (AF647) NHS ester (succinimidyl ester) (Invitrogen A37573) was dissolved into dimethyl sulfoxide (DMSO) at a final concentration of 1 mg ml−1. While stirring, we added 25 µg of the amine reactive dye to 1 mg of VLP and incubated the mixture for 1 h at room temperature. We removed excess dye from the solution with a Zeba Spin Desalting Column (Thermo Fisher Scientific) and buffer exchanged labelled VLPs into PBS and stored these at 4 °C. Labelled VLPs were used for confocal microscopy experiments within 12 h of labelling.
Ectopic expression experiments
cDNA encoding full length human VLDLR (GenBank NP_003374.3), human LDLR (GenBank AAP88892) and MXRA8 (clone ID: NM_032348.3) were obtained from GenScript. cDNA encoding human NRP2 (GenBank NM_201267.2)68 was provided by S. Whelan. Codon-optimized versions of the following H. sapiens VLDLR (GenBank NP_003374.3), H. sapiens ApoER2 isoform 1 (GenBank NM_004631.5), H. sapiens ApoER2 isoform 2 (GenBank NM_004631.5), M. musculus ApoER2 (GenBank XP_036019651), E. caballus VLDLR (GenBank XP_023483037), E. caballus ApoER2 (GenBank XP_023485552), S. vulgaris VLDLR (GenBank XM_014880599.1), S. vulgaris ApoER2 (GenBank XM_014870608.1), A. aegypti lipophorin receptor 1 (GenBank JN411069.1), A. albopictus lipophorin receptor 1 (GenBank JAC13440) and C. elegans VLDLR (GenBank NM_182223.6). For constructs containing a Flag tag, we used the SignalP server69 to predict signal peptidase processing sites and introduced sequence encoding a glutamic acid residue followed by a Flag tag (DYKDDDDK) and a short linker (GSG) at the N-terminus of each construct to monitor cell surface expression. We subcloned untagged or Flag-tagged versions of these constructs into the backbone of the lentiGuide-Puro vector (Addgene #52963)60. We packaged lentivirus in HEK 293T cells by co-transfecting this vector with psPAX2 (Addgene #12260), and a previously described pCAGGs vector expressing vesicular stomatitis virus (VSV) G70 in a ratio of 3:2:1 using Lipofectamine 3000 (Invitrogen). We exposed K562 cells to filtered (0.45 μm) supernatants containing lentivirus for 48 h. We selected transduced cell populations with puromycin (2 μg ml−1). For all Flag-tagged constructs, we used a FACS sorting step to select subpopulations of positive cells and confirmed construct cell surface expression using cell surface antibody staining (Extended Data Figs. 2b, 3a).
Generation of Fc fusion proteins and RAP
We subcloned the LBD of human VLDLR (residues 31–355, GenBank NP_003374.3), the LBD of human ApoER2 isoform 1 (46–334, GenBank NM_004631.5), the ectodomain of human MXRA8 (residues 20–337, GenBank NP_001269511.1), or the a1 subdomain of NRP2 (NRP2a1, residues 32–146, GenBank NM_201267.2) into a pVRC expression vector encoding the human IgG1 Fc provided by A. Schmidt71. We expressed these Fc fusion proteins in HEK 293T cells grown in suspension using linear polyethylenimine (PEI) according to the manufacturer’s protocol. We collected supernatants five days post-transfection and purified Fc fusion proteins with MabSelect SuRe LX protein A affinity resin (GE Healthcare) using the manufacturer’s protocol and further by size-exclusion chromatography using a Superdex 200 increase column. The Fc fusion proteins were stored in TBS containing 2 mM CaCl2.
VLDLRLBD–Fc fusion protein purification yields were poor when the protein was expressed alone. To increase VLDLRLBD–Fc fusion protein purification yields, we cloned full length human RAP (residues 1–353, including the signal sequence) (GenBank NP_002328) into the pCAGGs vector. Co-transfection of HEK 293T cells grown in suspension with vectors encoding VLDLRLBD–Fc and human RAP in a 1:1 ratio drastically improved expression yields. VLDLRLBD–Fc fusion protein and RAP co-eluted as a stable complex when purified using protein A affinity chromatography and subsequent size-exclusion chromatography on a Superdex 200 Increase column. We separated the VLDLRLBD–Fc from RAP by binding the complex to protein A resin and washing with 100 column volumes of 10 mM EDTA in TBS, followed by a wash with 50 column volumes of 10 mM EDTA and 500 mM NaCl in TBS. The washes were collected, buffer exchanged in TBS containing 2 mM CaCl2, and concentrated. RAP eluted as a single peak by size-exclusion chromatography using a Superdex 200 Increase column. The VLDLRLBD–Fc fusion protein was refolded on the column by washing with 100 column volumes of TBS containing 2 mM CaCl2 and eluted using the manufacturers protocol.
Prior to mouse studies and after size exclusion, the VLDLRLBD–Fcfusion protein was bound to a protein A resin and washed with 100 column volumes of 10 mM EDTA in TBS, followed by a wash with 100 column volumes of 0.5 M l-Arginine in TBS. The VLDLRLBD–Fc fusion protein was washed with 100 column volumes of TBS containing 2 mM CaCl2 and eluted. Endotoxin levels were <4 endotoxin units ml−1 for both the VLDLRLBD–Fc fusion protein and the control IgG used in mouse studies as quantified using a Pierce Chromogenic Endotoxin Quantification Kit (Thermo Fisher Scientific).
To generate Flag-tagged RAP (RAPFlag), we cloned RAP (residues 1–353) with a C-terminal Flag tag in place of the endoplasmic reticulum retention signal into the pCAGGS vector. We co-transfected HEK 293T cells in suspension with vectors encoding VLDLRLBD–Fc and human RAPFlag in a 1:1 ratio and purified the protein as described earlier.
To generate soluble VLDLR-LBD (sVLDLRLBD), we cloned VLDLR residues 31–355 with an N-terminal twin-Strep tag (WSHPQFEKGGGSGGGGSGGSAWSHPQFEK) followed by a Factor Xa cleavage site (IEGR) followed by a SGSG linker into the pHLsec vector64. We co-transfected HEK 293T cells grown in suspension with plasmids encoding sVLDLRLBD and RAP in a 1:1 ratio using PEI and by following the manufacturer’s protocol. We collected supernatants five days post-transfection and treated the supernatant with BioLock (Iba Lifesciences 2-0205-050), according to the manufacturer’s protocol, to sequester biotin from the media. We bound the sVLDLRLBD-RAP complex to Strep-Tactin XT Resin (IBA-Lifesciences 2-4030-010) and washed the column with 150 column volumes of 10 mM EDTA in TBS to remove RAP, followed by washing with 50 column volumes of 2 mM CaCl2 in TBS to refold sVLDLRLBD. We eluted the protein with buffer containing 50 mM biotin, 2 mM CaCl2 in TBS followed by dialysis in 2 mM CaCl2 in TBS. We removed the Twin-Strep-tag by adding Factor Xa protease (NEB P8010L) to the sVLDLRLBD solution in a 1:50 ratio for 3 h, at which point we inactivated the reaction by adding 1,5-dansyl-Glu-Gly-Argchloromethyl ketone, dihydrochloride Calbiochem (Sigma 251700-5MG) to a final concentration of 2 μM. We separated sVLDLRLBD from the cleaved Twin-Strep tag by size-exclusion chromatography on a preparation grade Superdex 200 16/600 column.
Entry blocking assays with Fc fusion proteins, anti-VLDLR antibody, and RAP with immortalized cell lines
We pre-incubated alphavirus RVPs with Fc fusion proteins or RAP in culture media containing 5 µg ml−1 polybrene for 30 min at 37 °C. We then added RVP/Fc fusion protein or RVP/RAP mixtures to cells. For blocking studies with SINV–SFV and SINV–CHIKV chimeras, we pre-mixed chimeras at an MOI of 1 with monoclonal antibodies anti-human VLDLR 1H10 (GeneTex GTX79552) or anti-HLA (BD Biosciences 560187) for 30 min at 37 °C, then added the mixture to Vero (CCL-81) cells and incubated for 1 h at 37 °C, and then replaced media. We visualized cells by fluorescence microscopy (using a LAXCO LMC4-FL8 microscope, Olympus UPLFNL Semi-Apo Phase objectives, with a 470 nm LED fluorescence lamp fluorescence filter set) or measured GFP expression by FACS using an iQue3 Screener PLUS (Intellicyt) with IntelliCyt ForeCyt Standard Edition version 8.1.7524 (Sartorius) software or a BD LSR II Flow Cytometer (BD Biosciences) with BD FACSDiva (BD Biosciences) software 30 h post infection for RVPs and 24 h post infection for SINV chimeras, after washing cells twice with PBS then fixing with PBS containing 2% (v/v) formalin. We used IntelliCyt ForeCyt Standard Edition version 8.1.7524 (Sartorius) or FlowJo (BD Biosciences) version 10.6.2 to analyze data. An example of the flow cytometry gating scheme used to quantify GFP-expressing RVP infection is provided in Extended Data Fig. 1b. We calculated relative infection as follows: Relative infection (%) = (%GFP positive cells in the presence of antibody or Fc fusion protein or RAP)/(%GFP-positive cells in the absence of antibody or Fc fusion or RAP) × 100%.
For plaque-neutralization assays with infectious SFV A774, the VLDLRLBD–Fc fusion protein or an isotype control IgG (C1A-H12, a previously described SARS-CoV-2 non-neutralizing IgG1 antibody)72 were serially diluted tenfold in PBS with a starting concentration of 100 μg ml−1. Fifty PFUs of SFV A774 were mixed with the serially diluted VLDLRLBD–Fc fusion protein, isotype control antibody, or PBS. After incubation at 37 °C for 1 h, the protein–virus mixtures were inoculated onto 12-well plates with a monolayer of Vero cells (which had been seeded the previous day). The cells were incubated at 37 °C with 5% CO2 for 1 h with gentle shaking every 15 min. After 1 h, 1 ml of overlay medium (DMEM, 2% (v/v) FBS, 0.8% (v/v) methyl cellulose, and 1% penicillin/streptomycin) was added onto each well. The plates were cultured at 37 °C with 5% CO2 for 2 d until clear plaques formed. The plates were fixed in 4% (v/v) formaldehyde solution for 2 h and stained with 1% (v/v) crystal violet. Plaques were counted. We calculated relative infection as follows: Relative infection (%) = (number of plaques in the presence of antibody or Fc fusion protein)/(number of plaques in the absence of antibody or Fc fusion) × 100%.
Cell surface Fc fusion protein binding assays
We transfected HEK 293T cells with pCAGGS alphavirus E3–E2–(6K/TF)–E1 expressor plasmids using Lipofectamine 3000 (Invitrogen) or an empty pCAGGS vector. We detached cells 48 h post-transfection with TrypLE Express (Thermo Fisher Scientific) and washed them in 50 mM Tris HCL pH 7.5, 150 mM NaCl, 2 mM CaCl2, 2% (w/v) bovine serum albumin (BSA) (binding buffer) followed by incubation in 50 mM Tris HCL pH 7.5, 150 mM NaCl, 2 mM CaCl2, 4% (w/v) BSA (blocking buffer). Cells were then incubated with increasing concentrations of MXRA8ect–Fc or VLDLRLBD–Fc fusion protein in binding buffer with 3% (v/v) goat serum for 1 h at 4 °C. We then washed cells three times in binding buffer and incubated them with a phycoerythrin (PE)-coupled goat anti-human F(ab’)2 fragment (Jackson ImmunoResearch, 109-116-098) at 1:200 dilution in binding buffer for 1 h at 4 °C. We washed cells three times with binding buffer, then twice with binding buffer without BSA, and fixed cells with 2% (v/v) formalin. We measured cell binding by monitoring PE-intensity and percent positivity by FACS. To control for non-specific cell surface binding, we subtracted the percent positive cells detected in staining experiments with untransfected cells incubated with the same Fc fusion proteins.
Confocal microscopy with labelled virus-like particles
We spun down 10 × 106 K562 cells transduced for ectopic expression of membrane proteins in a 15 ml conical tube at 1200 RPM for 3 min. We removed supernatant and treated the cells with a Heparinase I and III mixture (Sigma H3917) at 2 units ml−1 and Heparinase II (Sigma H8891) at 1 unit ml−1 for 1 h at 37 °C. Cells were washed and resuspended in 1.5 ml microcentrifuge tube at a concentration of 0.5 × 106 cells ml−1 in culture medium. Twenty-five micrograms of VLPs were added to 0.5 × 106 cells. For cells kept at 4 °C, after virus was added, 1.5 ml microcentrifuge tubes were immediately placed on ice and incubated for 15 min. Cells were then washed twice with PBS and kept at 4 °C before imaging. For the 37 °C condition, after adding virus to 1.5 ml microcentrifuge tubes, these were immediately placed at 37 °C and incubated for 15 min. Cells were then washed twice with PBS, and then kept at 4 °C. Just prior to imaging, 500 µl of WGA conjugated to Alexa Fluor 488 (WGA-AF488) (Invitrogen W11261) at 1 µg ml−1 was added to the cell mixture and incubated for 4 min on ice. Cells were then washed twice in PBS, resuspended in 80 µl of PBS, and placed in glass bottom microwell dishes (MatTek P35G-1.5-14-C) for immediate imaging. Samples were imaged with a Yokogawa CSU-W1 single disk (50 µm pinhole size) spinning disk confocal unitattached to either a fully motorized Nikon Ti or a Nikon Ti2 inverted microscope equipped with a Nikon linear-encoded motorized stage with a PI 250 mm range or a MadLabs 500 Z mm range piezo insert, an Andor Zyla 4.2 plus (6.5 µm photodiode size) sCMOS camera using a Nikon Plan Apo λS SR HP 100×C/1.45 Silicon DIC silicone immersion objective with Nikon Silicone oil. The final digital resolution of the image was 0.065 µm per pixel. Fluorescence from WGA-AF488 and VLPs conjugated to AF647 was collected by illuminating the sample with a solid-state directly modulated 488 nm diode 100 mW (at the fibre tip) laser line or a solid state, directly modulated 640 nm diode 100 mW (laser tip) laser line in a Toptica iChrome MLE laser launch, respectively. A hard-coated Semrock Di01-T405/488/568/647 multi-bandpass dichroic mirror was used for both channels. Signal from each channel was acquired sequentially with either a hard-coated Chroma ET525/36m or Chroma ET700/75m emission filters in a filter wheel placed within the scan unit, respectively. Z-stacks were set by determining the top and bottom of the cell, using WGA-AF488 fluorescence as a reference. The approximate volume was 20 µm, and the step size was set to 0.2 µm, using the piezo drive. Fluorescence from each fluorophore was acquired sequentially at each z-step of the confocal to improve the precision of the measurements. Nikon NIS-Elements Advanced Research (AR) 5.02 acquisition software was used to acquire the data, and the files were exported in ND2 file format. Figures were generated in Fiji73. A median filter of 1.0 pixels was applied to both channels (VLP and WGA). Gamma of 0.7 or 1.2 gamma was set for WGA and VLP filtered images, respectively, before adjusting brightness and contrast. Top views of stacks were created by using the (Stacks>Reslice>Top) function with an output spacing of 0.3 µm. 3D renderings were created by using the 3D projection function (Stacks>3D Project). The starting angle was set to 30° with 10° increments and interpolation selected to smooth the 3D rendering.
For VLP quantification of confocal images, 3D image analysis was performed using custom pipelines built in Arivis 4DFusion 3.4 analysis software. Viral particles were detected using a particle enhancement filter of 0.65 µm followed by a dilation morphology filter of diameter 0.52 µm (sphere shaped) and a Blob Finder segmentation filter set to 0/52 µm diameter, 3% probability threshold and split sensitivity of 70%. To segment cells, the signal from WGA-488 was first enhanced using the enhance edges filter within the membrane detection operation, selecting a membrane width of 0.6 µm and a gap size of 0.13 µm. A discrete gaussian filter of diameter 0.65 µm was applied to the resulting enhanced image. Finally, the membrane-based segmentation operation was used to segment the processed image to obtain the whole cell masks. The two compartments, cytoplasm and membrane, were created by eroding the cell mask by two pixels (cytoplasm) and by performing object math between the cell masks and the eroded cytoplasm masks, producing the membrane masks. The number of viruses in each compartment was then calculated by combining all masks.
Biolayer interferometry binding assays
We performed BLI experiments with an Octet RED96e (Sartorius) and analyzed data using ForteBio Data Analysis HT version 12.0.1.55 software. MXRA8ect–Fc, VLDLRLBD–Fc, or ApoER2LBDiso1–Fc were loaded onto Anti-Human IgG Fc Capture (AHC) Biosensors (Sartorius 18-5063) at 250 nM in kinetic buffer (TBS containing 2 mM CaCl2 and 0.1% (w/v) BSA) for 80 s. After a baseline measurement for 60 s in kinetic buffer, Fc fusion protein coated sensor-tips surfaces were incubated with RAP or transferrin at 100 µg ml−1 for 50 s, or kinetic buffer alone for 50 s. After an additional baseline measurement for 20 s in kinetic buffer, VLPs were associated for 300 s at 20 nM. We plotted total response nm change at the end of the 300 s association in Fig. 3b, and raw sensorgrams are provided in Extended Data Fig. 4c.
Cell surface staining of receptors and receptor orthologues
We incubated cells for 30 min at 4 °C in PBS containing 5% (v/v) goat serum (blocking buffer), prior to incubating them with an anti-human VLDLR monoclonal antibody 1H10 (GeneTex GTX79552) at 10 μg ml−1, anti-human ApoER2 (LRP8) antibody (clone 3H2) (Sigma WH0007804M1-100) at 10 μg ml−1, anti-human LDLR monoclonal antibody (R&D Systems MAB2148-100) at 10 μg ml−1, anti-human MXRA8 antibody (clone 2H2G12A) (MBL International W040-3) at 10 μg ml−1, or no antibody in PBS containing 2% (v/v) goat serum (binding buffer) for 1 h. Following incubation, we washed cells three times in binding buffer and then incubated cells for 30 min with a PE-conjugated donkey anti-mouse F(ab’)2 fragment (Jackson ImmunoResearch 715-116-150) at 1:200 dilution according to the manufacturer’s recommendedbinding buffer. We washed cells twice with binding buffer, fixed them with 2% (v/v) formalin, and detected cell surface receptor expression by FACS using an iQue3 Screener PLUS (Intellicyt) with ForeCyt (Sartorius) software.
For staining of cells expressing Flag-tagged receptors, we added an APC conjugated anti-DYKDDDDK (Flag) antibody (BioLegend 637307) at 1:200 dilution in binding buffer, according to the manufacturer’s recommendation. We washed cells three times with binding buffer, fixed them with 2% (v/v) formalin, and detected cell surface receptor expression by FACS. For staining using Flag-tagged RAP, we incubated cells with RAPFlag at 10 μg ml−1, or no protein in binding buffer for 30 min. Following incubation, we washed cells three times with binding buffer and added an APC conjugated anti-DYKDDDDK (BioLegend 637307) at 1:200 dilution and carried out the steps described above for antibody staining of Flag-tagged receptors.
Negative-stain electron microscopy
We collected negative-stain micrographs of VLPs at the Molecular Electron Microscopy Core Facility at Harvard Medical School. We buffer-exchanged VLPs into buffer containing 50 mM Tris HCl pH 7.4, 100 mM NaCl, 1 mM EDTA using a 100-kDa Amicon filter (Sigma). VLPs were adsorbed to glow-discharged Formvar carbon film grids (Electron Microscopy Sciences), rinsed twice with water, and stained with 1.5% uranyl formate. Multiple fields of view were inspected to confirm VLP integrity and the absence of associated lipoproteins. Representative micrographs shown in Extended Data Fig. 4b were collected using a Tecnai T12 (ThermoScientific) at 120 kV with a Gatan UltraScan 895 4k CCD camera.
Replication-competent virus replication kinetics assays
Transduced K562 cells (2.5 × 106) were spun in a 15 ml polypropylene conical tube at 1200 RPM for 5 mins; SFV (A774), SINV (DAK Ar Mg812), or EEEV (FL93-939) were used to inoculate cells at a multiplicity of infection (MOI) of 0.01. Cells were incubated with virus for 2 h in a 15 ml polypropylene conical tube in a CO2 incubator at 37 °C, washed three times with D-PBS (Lonza), and resuspended to a final concentration of 5 × 105 cells ml−1 with culture medium in T25 flasks (Corning). At indicated time points (0, 6, 12, 24 or 48 h), 500 μl of culture supernatants were collected for plaque assays, with 500 μl of fresh culture medium added back to the flask. Virus titres were then determined on Vero cells by plaque assay.
In vivo study
Mouse studies were performed in accordance with the NIH Guidance for the Care and Use of Laboratory Animals. The study protocol was approved by the University of Texas Medical Branch Institutional Animal Care and Use Committee under protocol 1708051. Pregnant mice were received by the dedicated animal research personnel at the University of Texas Medical Branch, who randomly assigned animals to one mouse per cage with no additional knowledge of the study design. Mice were fed a 19% protein diet (Teklad, 2919, Irradiated), had a 12 h light:dark cycle (0600–1800 h), and were housed in a facility maintained at a temperature range of 20 to 26 °C with a humidity range of 30 to 70%. Food and water were provided ad libitum. The offspring were too young to be randomly separated into different cages, and no further randomization was performed by study personnel. Pups were taken as mixed groups and were not sexed (that is, both sexes were used). All mouse manipulations were performed under anesthesia with isoflurane. Ten-day-old CD-1 mice (Charles River) were administered 125 μg VLDLRLBD–Fc fusion protein or IgG1 isotype control monoclonal antibody through the intraperitoneal route 6 h before intraperitoneal inoculation with 100 PFU or 1,000 PFU of SFV A774. Survival rate of mice was monitored daily.
Statistical analysis
Data were deemed statistically significant when P values were < 0.05 using version 9 of GraphPad Prism. Experiments were analysed by one-way or two-way ANOVA with multiple comparison correction or by log-rank (Mantel–Cox) test in GraphPad Prism. P values are indicated in each of the figure legends.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
All data that support the findings of this study are available within the Article and its Supplementary Information. Confocal microscopy images that support the findings of this study are available at https://omero.hms.harvard.edu/webclient/?show=project-8752. Any other relevant data are available from the corresponding author upon reasonable request. Source data are provided with this paper.
Code availability
Custom pipelines built in Arivis 4DFusion 3.4 analysis software used for this study are available at https://github.com/paulamonterollopis/Viral_Particle_on_Cells_Arivis.
References
Rose, P. P. et al. Natural resistance-associated macrophage protein is a cellular receptor for Sindbis virus in both insect and mammalian hosts. Cell Host Microbe 10, 97–104 (2011).
Zhang, R. et al. Mxra8 is a receptor for multiple arthritogenic alphaviruses. Nature 557, 570–574 (2018).
Ma, H. et al. LDLRAD3 is a receptor for Venezuelan equine encephalitis virus. Nature 588, 308–314 (2020).
Cheng, R. H. et al. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell 80, 621–630 (1995).
Paredes, A. M. et al. Three-dimensional structure of a membrane-containing virus. Proc. Natl Acad. Sci. USA 90, 9095–9099 (1993).
Paredes, A. M., Simon, M. N. & Brown, D. T. The mass of the Sindbis virus nucleocapsid suggests it has T = 4 icosahedral symmetry. Virology 187, 329–332 (1992).
Kuhn, R. J., Niesters, H. G., Hong, Z. & Strauss, J. H. Infectious RNA transcripts from Ross River virus cDNA clones and the construction and characterization of defined chimeras with Sindbis virus. Virology 182, 430–441 (1991).
Nimpf, J. & Schneider, W. J. From cholesterol transport to signal transduction: low density lipoprotein receptor, very low density lipoprotein receptor, and apolipoprotein E receptor-2. Biochim. Biophys. Acta 1529, 287–298 (2000).
Poirier, S., Mamarbachi, M., Chen, W. T., Lee, A. S. & Mayer, G. GRP94 regulates circulating cholesterol levels through blockade of PCSK9-induced LDLR degradation. Cell Rep. 13, 2064–2071 (2015).
Marceau, C. D. et al. Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature 535, 159–163 (2016).
Zhang, R. et al. A CRISPR screen defines a signal peptide processing pathway required by flaviviruses. Nature 535, 164–168 (2016).
Willnow, T. E., Armstrong, S. A., Hammer, R. E. & Herz, J. Functional expression of low density lipoprotein receptor-related protein is controlled by receptor-associated protein in vivo. Proc. Natl Acad. Sci. USA 92, 4537–4541 (1995).
Lozzio, C. B. & Lozzio, B. B. Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood 45, 321–334 (1975).
Dlugosz, P. & Nimpf, J. The reelin receptors apolipoprotein E receptor 2 (ApoER2) and VLDL receptor. Int. J. Mol. Sci. 19, 3090 (2018).
D’Arcangelo, G. et al. Reelin is a ligand for lipoprotein receptors. Neuron 24, 471–479 (1999).
D’Arcangelo, G. et al. A protein related to extracellular matrix proteins deleted in the mouse mutant reeler. Nature 374, 719–723 (1995).
Brandes, C. et al. Avian and murine LR8B and human apolipoprotein E receptor 2: differentially spliced products from corresponding genes. Genomics 42, 185–191 (1997).
Kim, D. H. et al. Human apolipoprotein E receptor 2. A novel lipoprotein receptor of the low density lipoprotein receptor family predominantly expressed in brain. J. Biol. Chem. 271, 8373–8380 (1996).
Clatworthy, A. E. et al. Expression and alternate splicing of apolipoprotein E receptor 2 in brain. Neuroscience 90, 903–911 (1999).
Lane-Donovan, C. & Herz, J. The ApoE receptors Vldlr and Apoer2 in central nervous system function and disease. J. Lipid Res. 58, 1036–1043 (2017).
Akahata, W. et al. A virus-like particle vaccine for epidemic Chikungunya virus protects nonhuman primates against infection. Nat. Med. 16, 334–338 (2010).
Ko, S. Y. et al. A virus-like particle vaccine prevents equine encephalitis virus infection in nonhuman primates. Sci. Transl. Med. 11, eaav3113 (2019).
Fazakerley, J. K. Semliki forest virus infection of laboratory mice: a model to study the pathogenesis of viral encephalitis. Arch. Virol. Suppl. 2004, 179–190 (2004).
Willems, W. R. et al. Semliki forest virus: cause of a fatal case of human encephalitis. Science 203, 1127–1129 (1979).
Lagomarsino, V. N. et al. Stem cell-derived neurons reflect features of protein networks, neuropathology, and cognitive outcome of their aged human donors. Neuron 109, 3402–3420 e3409 (2021).
Bradish, C. J., Allner, K. & Maber, H. B. The virulence of original and derived strains of Semliki forest virus for mice, guinea-pigs and rabbits. J. Gen. Virol. 12, 141–160 (1971).
Bradish, C. J. & Allner, K. The early responses of mice to respiratory or intraperitoneal infection by defined virulent and avirulent strains of Semliki forest virus. J. Gen. Virol. 15, 205–218 (1972).
Pattyn, S. R., De Vleesschauwer, L. & van der Groen, G. Replication of arboviruses in mouse organ cultures. II. Multiplication of virulent and avirulent Semliki Forest and western equine encephalitis viruses in mouse organ cultures. Arch. Virol. 49, 33–37 (1975).
Fleming, P. Age-dependent and strain-related differences of virulence of Semliki Forest virus in mice. J. Gen. Virol. 37, 93–105 (1977).
Woodward, C. G., Marshall, I. D. & Smith, H. Investigations of reasons for the avirulence of the A7 strain of Semliki Forest virus in adult mice. Br. J. Exp. Pathol. 58, 616–624 (1977).
Trommsdorff, M. et al. Reeler/Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell 97, 689–701 (1999).
Van Hoof, D., Rodenburg, K. W. & Van der Horst, D. J. Insect lipoprotein follows a transferrin-like recycling pathway that is mediated by the insect LDL receptor homologue. J. Cell Sci. 115, 4001–4012 (2002).
Van der Horst, D. J., Roosendaal, S. D. & Rodenburg, K. W. Circulatory lipid transport: lipoprotein assembly and function from an evolutionary perspective. Mol. Cell. Biochem. 326, 105–119 (2009).
Finkelshtein, D., Werman, A., Novick, D., Barak, S. & Rubinstein, M. LDL receptor and its family members serve as the cellular receptors for vesicular stomatitis virus. Proc. Natl Acad. Sci. USA 110, 7306–7311 (2013).
Ashrafi, K. et al. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421, 268–272 (2003).
Dieckmann, M., Dietrich, M. F. & Herz, J. Lipoprotein receptors-an evolutionarily ancient multifunctional receptor family. Biol. Chem. 391, 1341–1363 (2010).
Atkins, G. J., Sheahan, B. J. & Mooney, D. A. Pathogenicity of Semliki Forest virus for the rat central nervous system and primary rat neural cell cultures: possible implications for the pathogenesis of multiple sclerosis. Neuropathol. Appl. Neurobiol. 16, 57–68 (1990).
Johnson, R. T., McFarland, H. F. & Levy, S. E. Age-dependent resistance to viral encephalitis: studies of infections due to Sindbis virus in mice. J. Infect. Dis. 125, 257–262 (1972).
Hofer, F. et al. Members of the low density lipoprotein receptor family mediate cell entry of a minor-group common cold virus. Proc. Natl Acad. Sci. USA 91, 1839–1842 (1994).
Marlovits, T. C., Abrahamsberg, C. & Blaas, D. Very-low-density lipoprotein receptor fragment shed from HeLa cells inhibits human rhinovirus infection. J. Virol. 72, 10246–10250 (1998).
Bates, P., Young, J. A. & Varmus, H. E. A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell 74, 1043–1051 (1993).
Yamamoto, S. et al. Lipoprotein receptors redundantly participate in entry of Hepatitis C virus. PLoS Pathog. 12, e1005610 (2016).
Ujino, S. et al. Hepatitis C virus utilizes VLDLR as a novel entry pathway. Proc. Natl Acad. Sci. USA 113, 188–193 (2016).
Agnello, V., Abel, G., Elfahal, M., Knight, G. B. & Zhang, Q. X. Hepatitis C virus and other Flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl Acad. Sci. USA 96, 12766–12771 (1999).
Ganaie, S. S. et al. Lrp1 is a host entry factor for Rift Valley fever virus. Cell 184, 5163–5178.e5124 (2021).
Tao, L. et al. Sulfated glycosaminoglycans and low-density lipoprotein receptor contribute to Clostridium difficile toxin A entry into cells. Nat. Microbiol. 4, 1760–1769 (2019).
Demogines, A., Abraham, J., Choe, H., Farzan, M. & Sawyer, S. L. Dual host-virus arms races shape an essential housekeeping protein. PLoS Biol. 11, e1001571 (2013).
Gruszczyk, J. et al. Transferrin receptor 1 is a reticulocyte-specific receptor for Plasmodium vivax. Science 359, 48–55 (2018).
Li, W. et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014).
Saul, S. et al. Differences in processing determinants of nonstructural polyprotein and in the sequence of nonstructural protein 3 affect neurovirulence of Semliki Forest virus. J. Virol. 89, 11030–11045 (2015).
Finkbeiner, S. & Stevens, C. F. Applications of quantitative measurements for assessing glutamate neurotoxicity. Proc. Natl Acad. Sci. USA 85, 4071–4074 (1988).
Zhang, Y. et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 78, 785–798 (2013).
Vierbuchen, T. et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463, 1035–1041 (2010).
Maherali, N. et al. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell 3, 340–345 (2008).
Jemielity, S. et al. TIM-family proteins promote infection of multiple enveloped viruses through virion-associated phosphatidylserine. PLoS Pathog. 9, e1003232 (2013).
Bausch-Fluck, D. et al. A mass spectrometric-derived cell surface protein atlas. PLoS ONE 10, e0121314 (2015).
Almen, M. S., Nordstrom, K. J., Fredriksson, R. & Schioth, H. B. Mapping the human membrane proteome: a majority of the human membrane proteins can be classified according to function and evolutionary origin. BMC Biol. 7, 50 (2009).
da Cunha, J. P. Et al. Bioinformatics construction of the human cell surfaceome. Proc. Natl Acad. Sci. USA 106, 16752–16757 (2009).
Joung, J. et al. Genome-scale CRISPR–Cas9 knockout and transcriptional activation screening. Nat. Protoc. 12, 828–863 (2017).
Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014).
Doench, J. G. et al. Rational design of highly active sgRNAs for CRISPR–Cas9-mediated gene inactivation. Nat. Biotechnol. 32, 1262–1267 (2014).
Ran, F. A. et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389 (2013).
Saleh, M. C. et al. Antiviral immunity in Drosophila requires systemic RNA interference spread. Nature 458, 346–350 (2009).
Aricescu, A. R., Lu, W. & Jones, E. Y. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D 62, 1243–1250 (2006).
Erasmus, J. H. et al. Novel insect-specific Eilat virus-based chimeric vaccine candidates provide durable, mono- and multivalent, single-dose protection against lethal alphavirus challenge. J. Virol. 92, e01274-17 (2018).
Peng, J. & Gygi, S. P. Proteomics: the move to mixtures. J. Mass Spectrom. 36, 1083–1091 (2001).
Eng, J. K., McCormack, A. L. & Yates, J. R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass. Spectrom. 5, 976–989 (1994).
Raaben, M. et al. NRP2 and CD63 are host factors for Lujo virus cell entry. Cell Host Microbe 22, 688–696.e685 (2017).
Petersen, T. N., Brunak, S., von Heijne, G. & Nielsen, H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 (2011).
Radoshitzky, S. R. et al. Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature 446, 92–96 (2007).
Bajic, G. et al. Influenza antigen engineering focuses immune responses to a subdominant but broadly protective viral epitope. Cell Host Microbe 25, 827–835.e826 (2019).
Clark, S. A. et al. SARS-CoV-2 evolution in an immunocompromised host reveals shared neutralization escape mechanisms. Cell 184, 2605–2617.e2618 (2021).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014).
Acknowledgements
J.A. is a recipient of a William Randolph Hearst Foundation and Brigham and Women’s Hospital Young Investigator in Medicine Award, and a Burroughs Wellcome Fund Career Award for Medical Scientists. This work was also supported by a Harvard Milton Fund Award (J.A.), Vallee Scholar Award (J.A.), NIH grant T32 AI007061 (J.A.), NIH grant R24 AI120942 (S.C.W.), NIH grant T32 GM007753 (A.C., K.G.N. and D.V.N.), NIH grant R01 DK127257 (I.M.C.), Burroughs Wellcome Fund Pathogenesis Award (I.M.C.), and NIH T32 CA009216-40 (C.L.), and in part by a grant to Harvard Medical School from the Howard Hughes Medical Institute through the James H. Gilliam Fellowships for Advanced Study program (L.E.C.). The authors acknowledge the MicRoN (Microscopy Resources on the North Quad) Core at Harvard Medical School and the Molecular Electron Microscopy Core Facility at Harvard Medical School for their support and assistance in this work. Additionally, the authors thank A. Burdyniuk and M. Burdyniuk for support in building custom image analysis pipelines; and R. Tomaino from the Taplin Biological Mass Spectrometry Facility at Harvard Medical School for assistance with mass spectrometry of VLPs and data analysis.
Author information
Authors and Affiliations
Contributions
C.L. designed the sgRNA library and the RVP system and performed the CRISPR–Cas9 genetic screen and initial validation. L.E.C. generated cell lines, RVPs and recombinant proteins, and performed infectivity studies for validation with RVPs with assistance from S.A.C., A.C., K.G.N., D.V.N., H.L. and V.B. S.A.C. produced recombinant proteins, and generated cell lines, SINV chimeras and VLPs, and performed experiments with VLPs and SINV chimeras. S.A.C. additionally performed mass spectrometry experiments, BLI experiments and confocal microscopy experiments, the latter of which were performed with assistance from P.M.L. P.M.L. developed the imaging workflow and analysed confocal microscopy data with S.A.C. S.A.C., A.C., P.Y. and V.B. purified RVPs and VLPs for characterization, and A.C. performed negative-stain electron microscopy with VLPs. J.L., K.S.P. and S.C.W. designed and executed experiments with wild-type, replication-competent viruses including in vitro and in vivo studies. D.V.N. and I.M.C. provided mouse cortical neurons and assisted with RVP infectivity studies of mouse and human cortical neurons. H.L. and T.L.Y.-P. provided human iPS cell-derived neurons. I.S., A.A.A. and F.C. participated in study conceptualization or provided critical reagents. I.M.C., S.C.W. and J.A. acquired funding. J.A. wrote the original draft of the manuscript and all authors participated in reviewing and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review information
Nature thanks Laurie Silva and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Screening strategy, reporter virus particle system, and gating strategy.
a, Ross River (RRV) reporter virus particle (RVP) system. Cells are transfected with two plasmids. CD20 or GFP is included as a reporter downstream of the capsid (C) after a 2A peptide sequence. The arrow indicates a subgenomic promoter. b, SDS-PAGE gel of purified RVPs imaged with a stain-free imaging system. The experiment was performed twice independently, and a representative gel image is shown. c, Screening strategy. HEK 293T-Cas9 cells are first transduced with the guide (sgRNA) library using vesicular stomatitis virus (VSV) glycoprotein G pseudotyped lentiviruses and are then infected with RVPs expressing CD20. Infected cells are depleted using magnetic beads against CD20. Selection is repeated iteratively to improve the signal-to-noise ratio of the screen. Non-infected, CD20 negative cells are sequenced using next generation sequencing at the final step. See Methods for additional details. d, Coomassie-stained SDS-PAGE gel of purified virus-like particles (VLPs). The experiment was performed twice independently, and a representative gel image is shown. e, Flow cytometry gating strategy for quantification of GFP-expressing cells after RVP infection. K562 cells expressing human VLDLR (top panels) or wild-type (WT) K562 cells (bottom panels) were infected with GFP-expressing SFV RVPs. The percentage of cells falling within each gate is shown. The example is from an experiment shown in Fig. 4e. f, Flow cytometry gating strategy for detection of receptor cell surface staining. K562 cells overexpressing VLDLR (top panels) or WT K562 cells (bottom panels) were stained with RAPFLAG and a FLAG-APC antibody was used for detection. In the rightmost panel, the staining of each cell type is overlaid to allow for comparison. The example is from an experiment shown in Extended Data Fig. 3c. M: molecular weight marker. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 2 Knockout cell line validation and antibody blocking of SFV E2–E1-mediated entry into multiple cell lines.
a, Genotyping DNA gel (left panel) and anti-VLDLR (α-VLDLR) antibody cell surface staining of WT HEK 293T (middle panel) or HEK 293T VLDLR clonal knockout (K.O.) cells (right panel) as monitored by FACS. The experiment was performed at least twice independently, and a representative gel image is shown. b, Anti-VLDLR (α-VLDLR) cell surface staining of WT HEK 293T, HEK 293T VLDLR K.O., and HEK 293T VLDLR K.O. cells transiently transfected with cDNA encoding VLDLR-Flag (VLDLRFLAG) as monitored by FACS. c, α-VLDLR cell surface staining of the indicated cell types as monitored by FACS. d, The indicated cell types were infected with GFP-expressing SFV single-cycle RVPs in the presence or absence of a α-VLDLR or an anti-HLA control antibody (α-HLA) and infection was measured by FACS. Means ± standard deviation from two experiments performed in triplicate (n = 6) are shown. One-way ANOVA with Tukey’s multiple comparisons test, ****P < 0.0001 (d). For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 3 Immunostaining to monitor cell surface receptor expression.
a, Anti-FLAG (α-FLAG) and anti-MXRA8 (α-Mxra8), staining of WT K562 cells or K562 cells expressing the indicated constructs as monitored by FACS. b, Anti-ApoER2 (α-ApoER2) and anti-LDLR (α-LDLR) staining of the indicated cell types as monitored by FACS. c, RAPFLAG staining of WT K562 cells or K562 cells transduced with the indicated constructs as monitored by ɑ-FLAG-tag staining and FACS.
Extended Data Fig. 4 VLDLR and ApoER2 ligand binding domains directly bind alphavirus E2–E1 proteins.
a, Size exclusion chromatography traces of the indicated purified proteins. Insets are SDS-PAGE gels of the peak fraction. Molecular weight markers are indicated. Each experiment was performed at least twice, and representative traces are shown. b, Electron micrographs of negatively stained purified VLPs. Scale bar is 100 nm. The experiment was performed twice, and representative micrographs are shown. c, Sensorgrams for binding of the indicated alphavirus VLPs to tips coated with VLDLRLBD-Fc, ApoER2LBDiso1-Fc, or Mxra8ect-Fc fusion proteins as measured by biolayer interferometry. Fc fusion protein coated sensor-tips surfaces were incubated with RAP or transferrin, or kinetic buffer alone, and VLPs were associated followed by dissociation. The experiment was performed twice and representative results from one experiment are shown.
Extended Data Fig. 5 Role of VLDLR and ApoER2 in E2–E1-mediated cellular infection by divergent alphaviruses.
a, Wild-type (WT) or clonal VLDLR knockout (K.O.) HEK 293T cells were infected with GFP-expressing single-cycle alphavirus RVPs with relative infection measured by FACS. EEEV RVPs more efficiently entered VLDLR K.O. cells, which we suspect could be related to clonal variability, as the cell line was generated by clonal dilution. b, Vero cells were infected with GFP-expressing alphavirus single-cycle RVPs in the presence of the indicated antibodies with relative infection measured by FACS. c, Infection of WT or transduced K562 cells with GFP-expressing single-cycle RVPs. Cells were imaged by fluorescence microscopy. Scale bar is 100 μm. The experiment was performed twice, and representative images are shown. d, Infection of WT or transduced K562 cells with GFP-expressing single-cycle RVPs measured by FACS. NRP2 is a control membrane protein. e, K562 cells expressing VLDLR or ApoER2iso2 were infected with the indicated single-cycle RVPs in the presence of RAP, soluble VLDLR LBD (sVLDLRLBD), or a control protein (transferrin, Tf) with infection measured by FACS. f, WT or transduced K562 cells were infected with the indicated GFP-expressing single-cycle RVPs with infection measured by FACS. g, SFV A774 plaque reduction neutralization test with the indicated proteins performed on Vero cells. h, WT K562 cells or K562 cells transduced to express LDLRAD3 were infected with the indicated GFP-expressing single-cycle RVPs with infection measured by FACS. Means ± standard deviation from an experiment performed once in triplicate (n = 3) (a), or experiments performed twice in triplicate (n = 6) with similar results (b, d–h). One-way ANOVA with Tukey’s multiple comparisons test, ****P < 0.0001 (a, b, d–h). Two-way ANOVA with Šídák’s multiple comparison test, ****P < 0.0001 (g). Cell surface expression of constructs used in (c), (d), and (f) was confirmed with immunostaining (see Extended Data Fig. 3).
Extended Data Fig. 6 Ligand-binding domain sequence alignment and domain organization of ApoER2 constructs.
a, Sequence alignment of the Homo sapiens, Mus musculus, Equus caballus, and Sturnus vulgaris ApoER2 ligand binding domains. The LDLR class A (LA) repeats contained in each protein are shown in parentheses. The domain numbering is based on the human sequence shown. b, Schematic representation of the ectodomains of ApoER2 constructs used in this study. In mammals, exon regions encoding LA repeats 4-6 are almost exclusively spliced out, while the predominant avian isoforms retain these repeats14. Panel (a) was generated using ESPrit 3.074.
Extended Data Fig. 7 Representative confocal microscopy images for virus-like particle cell binding and internalization.
K562 cells transduced with human VLDLR, human ApoER2iso2, or human MXRA8 were incubated with fluorescently labeled VLPs at 4 °C or 37 °C and then imaged by live cell confocal microscopy. WGA: wheat germ agglutinin. Scale bar is 10 μm. The experiment was performed twice independently, and representative images are shown.
Extended Data Fig. 8 Workflow diagram of the 3-dimensional quantification of virus-like particle cell surface membrane binding and internalization.
a, 3D analysis of multi-colored stacks (pink, VLPs; green, cell membranes) using Arivis 4DFusion. Two custom-made pipelines were used to detect VLPs and cellular compartments. b, VLPs: left panel shows 3D rendering of VLP stacks, and right panel shows 3D rendering of detected VLPs. c, Cellular compartments: left panel shows 3D rendering of cellular membranes stacks; right, top panel shows 3D rendering of the detected cytoplasms (red) overlayed with an enhanced-membrane filter (white); right, bottom panel shows 3D rendering of the detected membranes (yellow). Objects obtained in each pipeline where combined to quantify the number of VLPs in each cellular compartment. d, Top: single plane representation of the detected objects, showing VLPs in the cytoplasm and the membrane. Bottom: 3D-view of the same cell. Related to Fig. 3c and 3d.
Extended Data Fig. 9 Effects of VLDLRLBD-Fc and RAP on E2–E1-mediated neuron infection and viral replication assays.
a, Infection of human neurons derived from induced pluripotent stem cell (iPSCs) with GFP-expressing SFV single-cycle RVPs in the presence of the indicated proteins. Cells were imaged by fluorescence microscopy. The experiment was performed twice with representative images shown. b, Quantification of single-cycle SFV RVP infection of human iPSC-derived neurons for the experiment shown in (a) using a live cell imaging system (see Methods for additional details) . c, Merged phase contrast and fluorescent microscopy for the experiment with mouse cortical neurons shown in Fig. 4a. Scale bars are 100 μm. Magnification is 20X. d, Merged phase contrast and fluorescent microscopy images for the experiment with human neurons shown in (a). Scale bars are 100 μm. Magnification is 10X. e, Viral replication curve for SFV, EEEV, and SINV strains in transduced K562 cells. Means ± standard deviation from two experiments done in triplicate (n = 6) with one-way ANOVA with Tukey’s multiple comparisons test, ****P < 0.0001 (b). Means ± standard deviation from two experiments done in triplicate (n = 6) with two-way ANOVA with Tukey’s multiple comparisons test, *P = 0.0233, ****P < 0.0001 (e).
Extended Data Fig. 10 Sequence alignment and domain organization of VLDLR constructs and summary of observed effects with alphavirus RVPs.
a, Sequence alignment of the Homo sapiens, Mus musculus, Equus caballus, Sturnus vulgaris, Aedes aegypti, Aedes albopictus, and C. elegans VLDLR ortholog ligand binding domains. The LDLR class A (LA) repeats contained in each protein are shown in parentheses. The domain numbering is based on the human sequence shown. b, Schematic representation of the ectodomains of VLDLR constructs used in this study. c, Summary of effects observed with GFP-expressing RVP infection of K562 cells transduced to express various VLDLR or ApoER2 orthologs derived from data shown in Extended Data Fig. 5d and Fig. 4e and 4f. +++: RVP infection with greater than 50% GFP positive cells achieved with overexpression. ++: RVP infection with 20–50% GFP positive cells achieved with overexpression. +: RVP infection with 5–20% GFP positive cells achieved with overexpression. +/-: RVP infection with less than 5% GFP positive cells of unclear biological significance. -: no enhancement. Panel (a) was generated using ESPrit 3.074.
Supplementary information
Supplementary Figure 1
Uncropped gels for the indicated Extended Data Figures.
Supplementary Table 1
List of genes encoding membrane-associated proteins targeted by the CRISPR–Cas9 library.
Supplementary Table 2
List of genes and scores from the CRISPR–Cas9 screen after MAGeCK analysis.
Supplementary Table 3
Results of mass spectrometry analysis of purified virus-like particles.
Rights and permissions
About this article
Cite this article
Clark, L.E., Clark, S.A., Lin, C. et al. VLDLR and ApoER2 are receptors for multiple alphaviruses. Nature 602, 475–480 (2022). https://doi.org/10.1038/s41586-021-04326-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-04326-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.