Abstract
The skin serves as a physical barrier and an immunological interface that protects the body from the external environment1,2,3. Aberrant activation of immune cells can induce common skin autoimmune diseases such as vitiligo, which are often characterized by bilateral symmetric lesions in certain anatomic regions of the body4,5,6. Understanding what orchestrates the activities of cutaneous immune cells at an organ level is necessary for the treatment of autoimmune diseases. Here we identify subsets of dermal fibroblasts that are responsible for driving patterned autoimmune activity, by using a robust mouse model of vitiligo that is based on the activation of endogenous auto-reactive CD8+ T cells that target epidermal melanocytes. Using a combination of single-cell analysis of skin samples from patients with vitiligo, cell-type-specific genetic knockouts and engraftment experiments, we find that among multiple interferon-γ (IFNγ)-responsive cell types in vitiligo-affected skin, dermal fibroblasts are uniquely required to recruit and activate CD8+ cytotoxic T cells through secreted chemokines. Anatomically distinct human dermal fibroblasts exhibit intrinsic differences in the expression of chemokines in response to IFNγ. In mouse models of vitiligo, regional IFNγ-resistant fibroblasts determine the autoimmune pattern of depigmentation in the skin. Our study identifies anatomically distinct fibroblasts with permissive or repressive IFNγ responses as the key determinant of body-level patterns of lesions in vitiligo, and highlights mesenchymal subpopulations as therapeutic targets for treating autoimmune diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The scRNA-seq data, mouse fibroblast RNA-seq data and human fibroblast RNA-seq data have been deposited in the Genome Sequence Archive (GSA) with accession number PRJCA006797. The code for the vitiligo progression model is available at https://github.com/hydrays/vitiligo. Source data are provided with this paper.
References
Larsen, S. B., Cowley, C. J. & Fuchs, E. Epithelial cells: liaisons of immunity. Curr. Opin. Immunol. 62, 45–53 (2019).
Rognoni, E. & Watt, F. M. Skin cell heterogeneity in development, wound healing, and cancer. Trends Cell Biol. 28, 709–722 (2018).
Afshar, M. & Gallo, R. L. Innate immune defense system of the skin. Vet. Dermatol. 24, 32–38 (2013).
Taieb, A. & Picardo, M. Vitiligo. N. Engl. J. Med. 360, 160–169 (2009).
Alikhan, A., Felsten, L. M., Daly, M. & Petronic-Rosic, V. Vitiligo: a comprehensive overview Part I. Introduction, epidemiology, quality of life, diagnosis, differential diagnosis, associations, histopathology, etiology, and work-up. J Am. Acad. Dermatol. 65, 473–491 (2011).
Picardo, M. et al. Vitiligo. Nat. Rev. Dis. Primers 1, 15011 (2015).
Ezzedine, K., Eleftheriadou, V., Whitton, M. & van Geel, N. Vitiligo. Lancet 386, 74–84 (2015).
Taieb, A. Intrinsic and extrinsic pathomechanisms in vitiligo. Pigment Cell Res. 13, 41–47 (2000).
Spritz, R. A. & Santorico, S. A. The genetic basis of vitiligo. J. Invest. Dermatol. 141, 265–273 (2021).
Frisoli, M. L., Essien, K. & Harris, J. E. Vitiligo: mechanisms of pathogenesis and treatment. Annu. Rev. Immunol. 38, 621–648 (2020).
Le Poole, I. C., van den Wijngaard, R. M., Westerhof, W. & Das, P. K. Presence of T cells and macrophages in inflammatory vitiligo skin parallels melanocyte disappearance. Am. J. Pathol. 148, 1219–1228 (1996).
Ogg, G. S., Rod Dunbar, P., Romero, P., Chen, J. L. & Cerundolo, V. High frequency of skin-homing melanocyte-specific cytotoxic T lymphocytes in autoimmune vitiligo. J. Exp. Med. 188, 1203–1208 (1998).
Van den Boorn, J. G. et al. Autoimmune destruction of skin melanocytes by perilesional T cells from vitiligo patients. J. Invest. Dermatol 129, 2220–2232 (2009).
Maeda, Y. et al. Detection of self-reactive CD8+ T cells with an anergic phenotype in healthy individuals. Science 346, 1536–1540 (2014).
Cheuk, S. et al. CD49a expression defines tissue-resident CD8+ T cells poised for cytotoxic function in human skin. Immunity 46, 287–300 (2017).
Iannella, G. et al. Vitiligo: pathogenesis, clinical variants and treatment approaches. Autoimmun. Rev. 15, 335–343 (2016).
Rodrigues, M. et al. Current and emerging treatments for vitiligo. J. Am. Acad. Dermatol. 77, 17–29 (2017).
Kovacs, S. O. Vitiligo. J. Am. Acad. Dermatol. 38, 647–666 (1998).
Gregg, R. K., Nichols, L., Chen, Y., Lu, B. & Engelhard, V. H. Mechanisms of spatial and temporal development of autoimmune vitiligo in tyrosinase-specific TCR transgenic mice. J. Immunol. 184, 1909–1917 (2010).
Harris, J. E. et al. A mouse model of vitiligo with focused epidermal depigmentation requires IFN-gamma for autoreactive CD8+ T-cell accumulation in the skin. J. Invest. Dermatol. 132, 1869–1876 (2012).
Chatterjee, S. et al. A quantitative increase in regulatory T cells controls development of vitiligo. J. Invest. Dermatol. 134, 1285–1294, (2014).
Yang, L. et al. Interferon-gamma inhibits melanogenesis and induces apoptosis in melanocytes: a pivotal role of CD8+ cytotoxic T lymphocytes in vitiligo. Acta Derm. Venereol. 95, 664–670 (2015).
Wang, S. Q. et al. Interferon-γ induces senescence in normal human melanocytes. PLoS One 9, e93232 (2014).
Tulic, M. K. et al. Innate lymphocyte-induced CXCR3B-mediated melanocyte apoptosis is a potential initiator of T-cell autoreactivity in vitiligo. Nat. Commun. 10, 2178 (2019).
Richmond, J. M. et al. Keratinocyte-derived chemokines orchestrate T-cell positioning in the epidermis during vitiligo and may serve as biomarkers of disease. J. Invest. Dermatol. 137, 350–358 (2017).
Biswas, K. B. et al. GPNMB is expressed in human epidermal keratinocytes but disappears in the vitiligo lesional skin. Sci. Rep. 10, 4930 (2020).
Boukhedouni, N. et al. Type-1 cytokines regulate matrix metalloprotease-9 production and E-cadherin disruption to promote melanocyte loss in vitiligo. JCI Insight 4, e133772 (2020).
Ganju, P. et al. Microbial community profiling shows dysbiosis in the lesional skin of vitiligo subjects. Sci Rep. 6, 18761 (2016).
Yuan, X. et al. The impact of NBUVB on microbial community profiling in the lesional skin of vitiligo subjects. Microb. Pathog. 140, 103943 (2020).
Lang, K. S. et al. HLA-A2 restricted, melanocyte-specific CD8+ T lymphocytes detected in vitiligo patients are related to disease activity and are predominantly directed against MelanA/MART1. J. Invest. Dermatol. 116, 891–897 (2001).
Namazi, M. R. Neurogenic dysregulation, oxidative stress, autoimmunity, and melanocytorrhagy in vitiligo: can they be interconnected? Pigment Cell Res. 20, 360–363 (2007).
Lotti, T., D’Erme, A. M. & Hercogova, J. The role of neuropeptides in the control of regional immunity. Clin. Dermatol. 32, 633–645 (2014).
Palermo, B. et al. Specific cytotoxic T lymphocyte responses against Melan-A/MART1, tyrosinase and gp100 in vitiligo by the use of major histocompatibility complex/peptide tetramers: the role of cellular immunity in the etiopathogenesis of vitiligo. J. Invest. Dermatol. 117, 326–332 (2001).
Mantovani, S. et al. Dominant TCR-α requirements for a self antigen recognition in humans. J. Immunol. 169, 6253–6260 (2002).
Byrne, K. T., Zhang, P., Steinberg, S. M. & Turk, M. Autoimmune vitiligo does not require the ongoing priming of naive CD8 T cells for disease progression or associated protection against melanoma. J. Immunol. 192, 1433–1439 (2014).
Mosenson, J. A. et al. Mutant HSP70 reverses autoimmune depigmentation in vitiligo. Sci. Transl. Med. 5, 174ra128 (2013).
Miao, X. et al. PD-L1 reverses depigmentation in Pmel-1 vitiligo mice by increasing the abundance of Tregs in the skin. Sci Rep. 8, 1605 (2018).
Riding, R. L., Richmond, J. M. & Harris, J. E. Mouse model for human vitiligo. Curr. Protoc. Immunol. 124, e63 (2019).
Richmond, J. M. et al. Resident memory and recirculating memory T cells cooperate to maintain disease in a mouse model of vitiligo. J. Invest. Dermatol. 139, 769–778 (2019).
Rashighi, M. et al. CXCL10 is critical for the progression and maintenance of depigmentation in a mouse model of vitiligo. Sci. Transl. Med. 6, 223ra223 (2014).
Yu, Z. et al. Hoxc-dependent mesenchymal niche heterogeneity drives regional hair follicle regeneration. Cell Stem Cell 23, 487–500 (2018).
Groom, J. R. & Luster, A. D. CXCR3 in T cell function. Exp. Cell. Res. 317, 620–631 (2011).
Chang, H. Y. et al. Diversity, topographic differentiation, and positional memory in human fibroblasts. Proc. Natl Acad. Sci. USA. 99, 12877–12882 (2002).
Paus, R., Ito, N., Takigawa, M. & Ito, T. The hair follicle and immune privilege. J. Investig. Dermatol. Symp. Proc. 8, 188–194 (2003).
Farber, E. M., Nickoloff, B. J., Recht, B. & Fraki, J. E. Stress, symmetry, and psoriasis: possible role of neuropeptides. J. Am. Acad. Dermatol. 14, 305–311 (1986).
Boehncke, W.-H. & Schön, M. P. Psoriasis. Lancet 386, 983–994 (2015).
Greb, J. E. et al. Psoriasis. Nat. Rev. Dis. Primers 2, 16082 (2016).
Gabrielli, A., Avvedimento, E. V. & Krieg, T. Scleroderma. N. Engl. J. Med. 360, 1989–2003, (2009).
Kaul, A. et al. Systemic lupus erythematosus. Nat. Rev. Dis. Primers 2, 16039 (2016).
Boyd, A. S. & Neldner, K. H. Lichen planus. J. Am. Acad. Dermatol. 25, 593–619 (1991).
Tang, Y., Harrington, A., Yang, X., Friesel, R. E. & Liaw, L. The contribution of the Tie2+ lineage to primitive and definitive hematopoietic cells. Genesis 48, 563–567 (2010).
Deng, L. et al. A novel mouse model of inflammatory bowel disease links mammalian target of rapamycin-dependent hyperproliferation of colonic epithelium to inflammation-associated tumorigenesis. Am. J. Pathol. 176, 952–967 (2010).
McCubbrey, A. L., Allison, K. C., Lee-Sherick, A. B., Jakubzick, C. V. & Janssen, W. J. Promoter specificity and efficacy in conditional and inducible transgenic targeting of lung macrophages. Front. Immunol. 8, 1618 (2017).
Le Gal, F. A. et al. Direct evidence to support the role of antigen-specific CD8+ T cells in melanoma-associated vitiligo. J. Invest. Dermatol. 117, 1464–1470 (2001).
Lo, J. A. et al. Epitope spreading toward wild-type melanocyte-lineage antigens rescues suboptimal immune checkpoint blockade responses. Sci. Transl. Med. 13, eabd8636 (2021).
Hua, C. et al. Association of vitiligo with tumor response in patients with metastatic melanoma treated with pembrolizumab. JAMA Dermatol. 152, 45–51 (2016).
Herzberg, B. & Fisher, D. E. Metastatic melanoma and immunotherapy. Clin. Immunol. 172, 105–110 (2016).
Nardin, C. et al. Vitiligo under anti-PD1 therapy is associated with increased survival in melanoma patients. J. Am. Acad. Dermatol. 82, 770–772 (2020).
Halprin, K. M. Epidermal "turnover time"–a re-examination. Br. J. Dermatol. 86, 14–19 (1972).
Gellatly, K. J. et al. scRNA-seq of human vitiligo reveals complex networks of subclinical immune activation and a role for CCR5 in Treg function. Sci. Transl. Med. 13, eabd8995 (2021).
Chen, T. et al. An RNA interference screen uncovers a new molecule in stem cell self-renewal and long-term regeneration. Nature 485, 104–108 (2012).
Lander, A. D. Morpheus unbound: reimagining the morphogen gradient. Cell 128, 245–256 (2007).
Chen, C. C. et al. Organ-level quorum sensing directs regeneration in hair stem cell populations. Cell 161, 277–290 (2015).
Acknowledgements
This work was supported by grants from the National Key R&D Program of China (2017YFA0103500), the National Basic Research Program of China 973 Programs (2014CB849602), National Natural Science Foundation of China (11671415) and the Beijing Municipal Natural Science Foundation (7172192). We thank the NIBS Animal Facility for their expert handling and care of mice; the NIBS Biological Resource Centre for FACS; the NIBS imaging facility for assistance with the confocal microscope experiment; the NIBS sequencing centre for sample processing of 10x genomics RNA-seq; V. Hearing for providing the DCT and TYR peptide sequences for in-house-made antibodies; F. Shao for help with importing Ifngr1 KO mice; and all members of the laboratory of T.C. for discussions and technical support.
Author information
Authors and Affiliations
Contributions
T.C. and Z.X. conceived the project, designed the experiments and wrote the manuscript. Z.X. and D.C. performed most of the in vivo experiments. Y.D. participated in the lentivirus injection experiment. K.J. performed some of the immunofluorescent staining experiment. Z.X., H.H. and J.W. conducted bioinformatics analysis of single-cell sequencing data. W. Wu collected fibroblasts from different body positions and participated in the cell culture experiment. D.C., H.H., J.W. and Z.X. conducted the bioinformatics analysis of the RNA-seq data. Y.H. developed the mathematical model. L.Z., W. Wang, Y.Y. and J.C. collected samples from patients with vitiligo and healthy donors. S.L., C.L. and J.C. collected clinical data from patients with vitiligo and carried out statistical analysis. J.S. provided the CD8 depletion antibody in the CD8+ T cell depletion experiment.
Corresponding authors
Ethics declarations
Competing interests
All authors declare no competing interests related to this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Comprehensive analysis of different cell populations from patients with vitiligo and healthy donors.
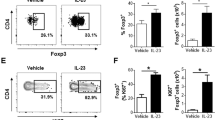
a, Representative images of the skin of patients with vitiligo showing characteristic bilateral symmetric lesion patterns in different body regions. b, Experimental workflow and representative FACS profiles to obtain single cells from human skin biopsies for single cell RNA-seq. c, Clinical information for all donors with the number of cells sequenced in single-cell analysis. d, t-SNE visualization of all collected cells showing eight main cell types with distinct clusters. e, Heat map analysis of differentially expressed genes in each cell type. f–h, violin plot (f), feature plot (g), and dot plot (h) analysis of signature genes for each cell type. i, Volcano plot and GO analysis of genes enriched in melanocytes from the M1 cluster compared to M2 cluster. j, T-SNE projection of T cells from patients with vitiligo and healthy donors. k, Volcano plot showing genes differentially expressed (>2-fold, t test p < 0.01) in each T cell sub-cluster. l, m, Representative immunofluorescent images (l) and quantification (m) of T cell subtype markers GZMA, ZNF683, CD40LG, and FOXP3 in T cells (CD3+) in the skin of patients with vitiligo. n, GO categories of genes enriched in each cell type from the skin of patients in the progressive state compared to healthy donor skin. o, Heat map analysis of 20 IFNγ response genes in eight cell types from the skin of patients in the progressive state and healthy donor skin. p, Violin plot showing the expression of IFNG in T cell subtypes. q, Violin plots showing IFNGR1 and IFNGR2 expression in different skin cell types. r, Representative immunofluorescent staining images, quantification (left and middle), and regression analysis (right) between density of CD8+ T cell and pSTAT1+ cell in the skin of patients with vitiligo. s, Representative immunofluorescent staining images of pSTAT1 signal in each cell type in the skin of patients with vitiligo. Scale bars, 50 µm (l, r, s). For exact p values, see Source Data. For statistics, p summary and sample sizes, see Methods.
Extended Data Fig. 2 Characterization of the melanoma–Treg-induced vitiligo mouse model.
a, Schematic diagram of the melanoma/Treg-induced vitiligo mouse model. First the dorsal skin in the right flank of 8–9-week old C57 mice was inoculated with B16F10 melanoma cells (Day 0), then CD4 depletion antibodies were injected on Days 4 and 10. The tumour was surgically removed on Day 12 to prevent interference of subsequent analysis. b, Representative hair coat images of control and vitiligo induced mice at Day 300 after the vitiligo induction procedure. At 4 weeks after induction, dorsal skin hair follicles close to the B16F10 tumour cell injection and surgical removal sites started to show depigmentation as a result of wounding-induced new hair growth. Then the depigmented hair follicles expanded and eventually rendered the whole dorsal hair coat depigmented at ~Day 300. c, Representative tail skin images of mice at Day 0, 45, 60, 90, 120, 240, 360, 480, 720 post melanoma/Treg-induced vitiligo induction procedure. Melanocytes in mouse dorsal skin are located in hair follicles but not in the epidermis; only in mouse tail skin are melanocytes located in both the hair follicle and epidermis similar to human skin. Skin epidermis depigmentation is the defining feature of vitiligo pathology. Therefore, we only used mouse tail skin for vitiligo analysis. In tail skin, the depigmentation was initially patchy and then progressed to eventually cover the entire epidermal surface, which did not recover even more than a year later. d, Representative whole-mount immunofluorescent staining and density plot images of DCT+ melanocytes and CD8+ T cells in mouse tail skin epidermis at Day 0, 19, 26, 33 after the vitiligo induction procedure. Prior to vitiligo induction, very few if any CD8+ T cells could be detected in epidermis. Starting from Day 19 after the vitiligo induction procedure, CD8+ T cells infiltration and small regions of melanocyte loss could be observed. Note, melanocytes loss only occurred in regions where CD8+ T cells locally aggregated into clusters; as the CD8+ T cells clusters continuously expanded so did the regions of melanocyte loss correspondingly. e, FACS quantifications of CD45+CD3+CD8+ T cells and CD117+ melanocyte in mice with or without vitiligo induction confirmed the loss of melanocytes and enrichment of CD8+ T cells after vitiligo induction. f, Representative whole-mount immunofluorescent staining images and quantifications of DCT+ melanocytes and CD8+ T cells in mouse tail skin epidermis with combined or individual B16F10 inoculation and CD4 depletion antibody treatment, showing the melanoma/Treg-induced vitiligo model requires both B16F10 inoculation and CD4 depletion antibody injection. g, Schematic diagram, representative whole-mount immunofluorescent staining images and quantification of DCT+ melanocytes and CD8+ T cells in the skin epidermis of melanoma/Treg-induced vitiligo model with or without CD8 depletion antibody treatment, showing CD8+ T cells are responsible for melanocyte loss in the melanoma/Treg-induced vitiligo mouse model. h, i, Representative whole-mount images (h) and FACS profiles (i) with corresponding quantification of the percentage of CD8+ T cells that express CD3 or CD11c in mouse epidermis, showing majority of the CD8+ cells are CD3+ T cells, but not CD11c+ dendritic cells. j, Representative whole-mount immunofluorescent staining images and density plot images of CD8+ T cells and DCT+ melanocytes in tail skin epidermis at Day 19, 26, 33 after vitiligo induction procedure. Note the continuously expanding CD8+ T cell clusters in skin epidermis. k, Representative whole-mount immunofluorescent staining images and quantifications of Ki67+, CD3+ T cells, and DCT+ melanocytes in tail skin epidermis at Day 33 after vitiligo induction procedure. Enlarged image on the left represents border of lesion skin, the right one represents lesion region. Quantification showed the proliferation rates of CD8+ T cells at the border versus inside the clusters are equivalent; and percentage of Ki67+ cells in melanocytes, T cells and keratinocytes showed majority of the proliferating cells in skin epidermis are keratinocytes. These data indicated that the continuous expansion of CD8+ T cell clusters mainly results from skin-infiltrated CD8+ T cells being actively recruited into the border regions. Scale bars, 500 µm (c, d, f, g, j), 100 µm (h, k). For exact p values, see Source Data. For statistics, p summary and sample sizes, see Methods.
Extended Data Fig. 3 The melanoma–Treg-induced vitiligo mouse model recapitulates hallmarks of human vitiligo.
a, UMAP projection of skin immune cells from patients with vitiligo and healthy donors. We identified 5 major clusters in immune cells, including two adaptive immune cell clusters (CD4+ T cell and CD8+ T cell) and three innate immune cell clusters (Langerhans cell, macrophage, dendritic cell 1/2). Other immune cells of very low abundance in human skin, such as mast cells and γδ T cells, could also be observed in our single-cell RNA-seq data, but owing to their limited number these cell types were not analysed individually. b–e, Heat map (b), dot plot (c), violin plot (d) and feature plot (e) analysis of signature genes in each immune cell subtype. f, Quantification and comparison of five immune cell types between progressive-state patients with vitiligo and healthy donors by scRNA-seq analysis. g, Representative FACS profiles analysing immune cell types in mice vitiligo skin, including epidermal TCRγδhigh and TCRγδlow γδ T cell present in mouse. h, i, Quantification and comparison of immune cell types between vitiligo (at Day 33) and control mice. Data in f-g showed the predominantly enriched immune cell type is CD8+ T cell in both patients with vitiligo and the melanoma/Treg-induced vitiligo mouse model, whereas the other immune cell types did not show significant difference. j, Representative whole-mount immunofluorescent staining images of CD4+ T cells in WT skin, and Tregs in Foxp3Cre;Ai14 skin, indicating at Day 33 of vitiligo analysis, the CD4+ T cell population have recovered from systematical CD4+ T cell depletion. k, Heat map analysis of cytotoxicity-related genes expressed in skin CD8+ T cells of patients with vitiligo and vitiligo-induced mice, indicating that in molecular level, this melanoma/Treg-induced vitiligo mouse model elicits endogenous auto-reactive CD8+ cytotoxic T cells similar to human vitiligo. Ctl: control, Vit: vitiligo. Scale bars, 200 µm (k). For exact p values, see Source Data. For statistics, p summary and sample sizes, see Methods.
Extended Data Fig. 4 Analysis of wild-type and Ifngr1 KO mice with three different vitiligo induction methods.
a–j, Analysis of WT and Ifngr1 KO mice with melanoma/Treg-induced vitiligo model. a, Schematic diagram. b, Representative tail skin images of WT and Ifngr1 KO mice at Day 60 after vitiligo induction. c, d, Representative whole-mount images (c) and quantification (d) of tail skin epidermal melanocytes and CD8+ T cells at Day 33 after vitiligo induction. e, f, Representative FACS profile of CD117+ melanocytes and CD3+CD8+ T cells (e), and quantification of CD117+ melanocytes (f) in the tail skin epidermis at Day 33 after vitiligo induction. g, Whole-mount views of scales in tail skin epidermis (dotted line) in control and vitiligo mice. This is the unit area we used to quantify melanocyte or CD8+ T cell density in wholemount analysis throughout this paper. h. Scatter plots and correlation analysis of melanocyte number versus CD8+ T cell number in each scale of WT and Ifngr1 KO tail skin at Day 33 after vitiligo induction. i, Representative whole-mount images of DCT+ melanocytes in tail skin epidermis of WT and Ifngr1 KO mice at 4 months after vitiligo induction. j, QPCR analysis of GZMA, GZMB, PRF1, and CCL5 expression in skin CD8+ T cells isolated from WT and Ifngr1 KO mice at Day 33 after vitiligo induction. k–p, Analysis of WT and Ifngr1 KO mice with Pmel transgenic spontaneous vitiligo mouse model. k, Schematic diagram. l–m, Representative tail skin images (l, P70) and whole-mount immunofluorescent staining images (m, P42) of Pmel;WT and Pmel; Ifngr1 KO mice. n, o, Representative FACS profiles (n) and quantification (o) of CD117+ melanocytes and CD45+CD3+CD8+ T cells in tail skin epidermis of WT, Ifngr1 KO, Pmel;WT and Pmel;Ifngr1 KO mice at P42. p, Scatter plots and correlation analysis of melanocyte number versus CD8+ T cell number in each scale in Pmel;WT and Pmel;Ifngr1 KO tail skin at P42. q–x, Analysis of WT and Ifngr1 KO mice with adoptive T cell transfer-based vitiligo mouse model. q, Schematic diagram. r, Quantification of CD117+ melanocytes and CD45+CD3+CD8+ T cells in tail skin epidermis of WT mice after γ irradiation, with or without WT Pmel T cell transfer or AVV-hPMEL intraperitoneal injection. These data indicate sub-lethal irradiation alone, irradiation with hPMEL-AAV alone, or irradiation with Pmel CD8+ T cell transfer alone did not result in melanocyte loss or CD8+ T cell infiltration in WT tail skin epidermis. s, Representative immunofluorescent staining images and FACS quantification of epidermal melanocyte of mice post γ-irradiation 26 days with controls, indicating after 5 Gy γ irradiation leads to ~2-fold increase of melanocyte number in tail skin epidermis compared to untreated mice. t, Representative FACS profiles and quantification of CD3+CD8+VB13+ Pmel T cells in spleen or skin of WT and Ifngr1 KO mice at Day 26 with or without adoptive transfer-based vitiligo model induction. These data showed although both WT and Ifngr1 KO host mice contained the same number of transferred WT TCR VB13+ Pmel T cells in the spleen, only WT host mice exhibit robust VB13+ Pmel T cell infiltration in skin, whereas the Ifngr1 KO host mice did not. u–v, Representative tail skin images (u, Day 60) and whole-mount immunofluorescent images (v, Day 26) of WT and Ifngr1 KO mice post vitiligo model-induction. Scale bars, 500 µm. w, Representative FACS profiles and quantification of epidermal CD117+ melanocytes in WT and Ifngr1 KO mice at Day 26 post vitiligo model-induction. x, Scatter plots and correlation analysis of melanocyte number versus CD8+ T cell number in each scale in WT and Ifngr1 KO mice at Day 26 post vitiligo model-induction. Scale bars, 500 µm (b, c, g, i, l, m, s, u, v). For exact p values, see Source Data. For statistics, p summary and sample sizes, see Methods.
Extended Data Fig. 5 IFNγ-responsive skin stromal cells are required for vitiligo progression.
a–c, Analysis of six main immune cell types in tail skin of WT and Ifngr1 KO mice with different vitiligo induction methods. a, Day 33 after melanoma/Treg-induced vitiligo model induction. b, P42 of Pmel;WT and Pmel;Ifngr1 KO mice. c, Day 26 after adoptive transfer-based vitiligo model-induction. d, Timeline of the graft and vitiligo induction assay. Representative whole-mount immunofluorescent staining images of grafted WT tail skin epidermis on WT host at Day 0 and Day 21 with or without vitiligo induction were showed. These data indicate tail skin did not spontaneously develop vitiligo after the graft procedure alone; only after vitiligo induction in the host mice did the grafted skin develop vitiligo as indicated by melanocyte loss and large amounts of CD8+ T cells infiltration. e, f, Lineage tracing experiment of CD8+ T cells: e, Schematic diagram WT tail skin graft on CD4Cre;mTmG host and representative immunofluorescent staining images of CD8+ T cells in the grafted skin epidermis after vitiligo induction; f, FACS analysis and quantification of spleen cells from CD4Cre;Ai6 mouse to determine GFP labelling efficiency in CD8+ T cells. Cells were pre-gated on CD45+ live singlets, indicating that the infiltrated CD8+ T cells were derived from the host mice rather than the grafted skin. These data indicate the graft infiltrated CD8+ T cells were derived from the host mice rather than the grafted skin. g, Quantification of melanocytes and CD8+ T cells number in grafted skin without vitiligo model induction in host mice. Donor skin pairs grafted onto the same host mouse are linked by lines. h, Representative immunofluorescent staining of the junction region between grafted C57 tail skin and host dorsal skin of a membrane-Tomato (mT) transgenic mouse. After full-thickness C57 tail skin was grafted onto membrane-tdTomato (mT)-expressing host mice (Rosa-mT), in which all host cells are genetically labelled as mT+, host derived dermal cells indeed invaded into the grafted skin dermis. But neither K14+ keratinocytes nor DCT+ melanocytes migrated from the host to grafted skin. i, Representative immunofluorescent staining image of pSTAT1 signal in immune cell (CD45+), smooth muscle cell (a-SMA+), Langerhans cells (Langerin+) of grafted Ifngr1 KO tail skin on WT host after vitiligo induction on a host mouse. Enlarged image on the right represents Langerin+ cells in the epidermis. Scale bars, 50 µm (d, e, h, i). For exact p values, see Source Data. For statistics, p summary and sample sizes, see Methods.
Extended Data Fig. 6 IFNγ-responsive skin fibroblasts are required for driving vitiligo pathogenesis.
a, Schematic diagram and representative FACS profiles of skin cells isolation. Detailed gating strategies are described in method section. QPCR analyses of cell-type-specific signature genes in FACS-isolated populations include KRT14 for keratinocytes, DCT for melanocytes, CD45 for immune cells, PDGFRA for fibroblasts, and CD31 for endothelial cells respectively. QPCR validation of IFNGR1 knockout used FACS-purified endothelial cells, keratinocytes, immune cells, melanocytes, fibroblasts and CD8+ T cells from TekCre;IFNGR1fl/fl, K14Cre;IFNGR1fl/fl, Csf1rCre;IFNGR1fl/fl, TyrCreER;IFNGR1fl/fl, CD4Cre;IFNGR1fl/fl, and PdgfraCreER;IFNGR1fl/fl mice, all with WT mice as controls. b, Representative whole-mount immunofluorescent staining images of melanocytes and CD8+ T cells in control and six cell-type-specific conditional knockout lines at Day 33 after vitiligo induction. c, Quantification of skin CD8+ T cells in WT and six cell-type-specific knockout lines at Day 33 after vitiligo induction procedure based on wholemount staining. d, e, Representative FACS profiles (d) and quantification (e) of CD117+ epidermal melanocytes and CD3+CD8+ T cells in control and six cell-type-specific conditional knockout lines post vitiligo induction. f, Schematic diagram of melanoma/Treg-induced vitiligo procedure and representative tail skin images (Day 60) of WT and PdgfraCreER;IFNGR1fl/fl cKO mice after vitiligo induction. Vit. induc.: Vitiligo induction. Scale bars, 500 µm (b, f). For exact p values, see Source Data. For statistics, p summary and sample sizes, see Methods.
Extended Data Fig. 7 IFNγ-responsive fibroblasts are both necessary and sufficient to mediate local recruitment of CD8+ T cells through secreted chemokines.
a–c, Analysis of WT and PdgfraCreER;IFNGR1fl/fl cKO mice with Pmel transgenic spontaneous vitiligo mouse model. a, Schematic diagram and representative tail skin images (P70) and representative wholemount images (P42) of Pmel;WT and Pmel;PdgfraCreER;IFNGR1fl/fl cKO mice. b, Scatter plots and correlation analysis of melanocyte number versus CD8+ T cell number in each scale from Pmel;WT and Pmel;PdgfraCreER;IFNGR1fl/fl cKO mice at P42. c, Representative FACS profiles and quantification of tail skin epidermal CD117+ melanocytes and CD3+CD8+ T cells in WT, PdgfraCreER;IFNGR1fl/fl cKO, Pmel;WT and Pmel;PdgfraCreER;IFNGR1fl/fl cKO mice at P42. d–g, Analysis of WT and PdgfraCreER;IFNGR1fl/fl cKO mice with adoptive transfer-based vitiligo model. d, Representative FACS profiles and quantification of CD3+CD8+VB13+ Pmel T cells in spleen or tail skin of WT and PdgfraCreER;IFNGR1fl/fl cKO mice at Day 26 with or without adoptive transfer-based vitiligo model induction. e, Schematic diagram and representative tail skin images (Day 60) and representative wholemount immunofluorescent staining images (Day 26) of WT and PdgfraCreER;IFNGR1fl/fl cKO mice after vitiligo induction. f, Scatter plots and correlation analysis of melanocyte number versus CD8+ T cell number in each scale WT and PdgfraCreER;IFNGR1fl/fl cKO mice at Day 26 after vitiligo induction. g, Representative FACS profiles and quantification of tail epidermal CD117+ melanocytes, and quantification of tail epidermal CD117+ melanocytes and CD3+CD8+ T cells of WT and PdgfraCreER; IFNGR1fl/fl cKO mice at Day 26. h, Section immunofluorescent analysis of intradermally injected RFP-labelled fibroblast. Note injected RFP+ fibroblasts located in lower dermis. i, Quantification of T cell transwell migration assay showing T cells migration ratio using control medium without fibroblast, corresponding to Fig. 3d. j, Analysis of human and mouse vitiligo fibroblasts up regulated genes. Left, heat map showing differential expressed genes in tail skin fibroblasts from WT control, WT vitiligo-induced, and Ifngr1 KO vitiligo-induced mice. Middle, volcano plot of differentially expressed genes in human fibroblasts from progressive-state patients with vitiligo compared with healthy donors. Red dots denote genes >1.5 fold upregulated (t test p < 0.01) in fibroblasts from progressive-state patients with vitiligo. Right, heat map analysis of the 28 common upregulated secreted factors in fibroblasts from patients with vitiligo and mice. k, Quantification of migration ratio from the T cell transwell migration assay with various chemokines. l, Representative immunofluorescent staining images of CXCL9 and CXCL10 signal with T cells (CD3+) in the skin of patients with vitiligo. m, n, Representative immunofluorescent staining images of CXCL9 (m) and CXCL10 (n) signals in melanocytes (DCT+), fibroblasts (PDGFRA+), endothelial cells (CD31+), smooth muscle cells (a-SMA+), keratinocytes (K14+), mononuclear phagocytes (CD11c+), Langerhans cells (Langerin+), and T cells (CD3+) in the skin of patients with vitiligo. Scale bars, 500 µm (a, e, l), 50 µm (h), 100 µm (m, n). For exact p values, see Source Data. For statistics, p summary and sample sizes, see Methods.
Extended Data Fig. 8 Fibroblasts directly affect CD8+ T cells through the CXCL9/CXCL10–CXCR3 axis in vitiligo.
a, Schematic diagram of the experimental procedure of lentivirus-infected cell type analysis. FACS profiles and quantification showing majority of lentivirus-infected cell population in the skin is fibroblast. b, QPCR analysis validated high knockdown efficiency of shRNAs targeting CXCL9/CXCL10 in vitro. c, Top, experimental design of in vivo knockdown efficiency detection in different dermal cell types. Bottom, qPCR analysis in FACS-isolated virus-infected (RFP+) fibroblasts, endothelial cells, and immune cells, indicating CXCL9 and CXCL10 expression were specifically silenced in virus-infected fibroblasts, not in other cell types. d, Representative whole-mount images showing dermis and overlying epidermis with injection of indicated shRNA-expressing lentivirus. Box-whisker plots of epidermal melanocyte number in each scale of mice tail skin injected with the indicated shRNAs. e, FACS quantification of immune cell types in tail skin of WT and PdgfraCreER;IFNGR1fl/fl cKO mice with three different vitiligo induction methods. f, Feature plot showing CXCR3 expression pattern and quantification of CXCR3 in all cell clusters from patients with vitiligo and healthy donors. g, Representative FACS profiles and quantification of CXCR3+ cells composition in vitiligo-induced mice (at Day 33). Scale bars, 500 μm (d). For exact p values, see Source Data. For statistics, p summary and sample sizes, see Methods.
Extended Data Fig. 9 Anatomically distinct dermal fibroblasts have intrinsic differences in the IFNγ response.
a, Heat map of upregulated genes in human dermal fibroblasts from different anatomical regions after IFNγ treatment (> 2-fold, t test p < 0.01). b, HOX expression pattern (based on RNA-seq results) in human dermal fibroblasts from eight different anatomic regions with or without IFNγ treatment during in vitro culture. c, QPCR analysis of HOXB8, HOXC8, HOXB13, and HOXD11 in human dermal fibroblasts from eight anatomic regions. d, JAK1, JAK2, IFNGR1, and STAT1 expression pattern (based on RNA-seq results) in human dermal fibroblasts from eight different anatomic regions with or without IFNγ treatment. e, QPCR analysis of TNFSF10, IL6, and CTSH in human dermal fibroblasts from eight anatomically distinct regions after IFNγ treatment. f, Left, representative whole-mount images and quantification of hair follicle density in the dorsal and paw dorsal skin of mice at P21. Right, representative section image and quantification of skin thickness in the dorsal and paw dorsal skin of mice at P60. g, Representative immunofluorescent images, and quantification of hair follicle located melanocytes in ventral and paw ventral skin of control mice and vitiligo mice (at Day 300). h, FACS analysis and quantification of skin CD3+CD8+ T cells and CD117+ melanocytes from paw dorsal, paw ventral, dorsal, and ventral regions at Day 300 after vitiligo induction. i, Schematic diagram of fibroblast isolation and qPCR analysis of CXCL9 and CXCL10 in fibroblasts from four distinct regions of long-term vitiligo mice. j, Quantification of migration ratio using medium without fibroblast, corresponding to Fig. 4g. k, Relationship between melanocyte number and infiltrated CD8+ T cell number in each scale after vitiligo induction based on quantification data in Extended Data Fig. 4h. Scale bars, 100 µm (f), 50 μm (g). For exact p values, see Source Data. For statistics, p summary and sample sizes, see Methods.
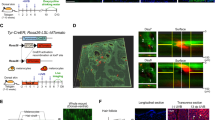
Extended Data Fig. 10 Mathematical modelling reveals that fibroblasts direct collective CD8+ T cell local activity.
a, The mathematic model developed to predict local CD8+ T cell recruitment and clonal expansion behaviour. The model is a 3D square lattice with two layers. The upper epidermal layer contains T cells and melanocytes. The lower dermal layer contains fibroblasts with different chemotactic abilities. The CD8+ T cell population in skin is considered to be a decentralized system. Each CD8+ T cell is equipped with the means of sensing a change in density. Over time, the behaviour of each cell changes according to its state and the states of its neighbouring cells and the surrounding signals. The collective pattern can be globally modulated by changing the parameters governing local cell-cell interactions. Once the T cell surrounding a melanocyte exceeds a threshold number, melanocyte death and IFNγ secretion occur. The IFNγ signal induce neighbouring fibroblast chemotactic effect to recruit nearby CD8+ T cells, reaching the local CD8+ T cell density threshold for adjacent melanocyte cytotoxicity and IFNγ secretion. This positive feedback loop between CD8+ T cells and fibroblasts could ensure T cell clonal expansion and vitiligo progression. b, The mathematic model predicts the expansion process of a single T cell clone. Five representative time points from T0 to T80 show the initial T cell cluster state and the subsequent T cell expansion process over 80 time units. In model 1 with normal chemotactic fibroblasts, full spectrum of T cell cluster formation and expansion patterns observed in WT vitiligo mouse model were reproduced (first row, fibroblasts with normal chemotactic effect, also shown in Supplementary Videos 1, 2). In model 2, in which the fibroblasts were incapable of chemotaxis, we obtained patterns observed in PdgfraCreER;IFNGR1fl/fl cKO mice. In this model, although T cells could still randomly aggregate in the epidermis, they failed to propagate this effect and recruit more T cells to the initial site (second row, fibroblasts with no chemotactic effect, also shown in Supplementary Videos 3, 4). In model 3, in which the chemotactic effect of fibroblasts was turned down to 1/2 of the normal value, random CD8+ T cell aggregates only recruited a limited number of T cells, resulting in slow T cell cluster expansion and melanocyte loss in one or two directions (third row, fibroblasts have 1/2 normal chemotactic effect, also shown in Supplementary Videos 5, 6). c, The mathematic model predicts large-scale T cell clone expansion over the long term. Four representative time points from T0 to T450 show the initial state and the subsequent T cell cluster expansion process over 450 time units. CD8+ T cells in the normal chemotaxis model efficiently coordinate so as to achieve clonal expansion and melanocyte clearance. CD8+ T cells in the no chemotaxis model fail to undergo clonal expansion and melanocyte death is detected. CD8+ T cells in the weak chemotaxis model (migration ability decreases to 1/2) generate small T cell clones and expand slowly. d, The mathematic model predicts the T cell clone distribution pattern under regional variant fibroblasts with different levels of chemotactic effect to T cell. In this model, the fibroblasts marked in blue have a normal chemotactic effect, and those marked in green have a weak chemotactic effect. In this region, T cell chemotaxis (migration ability) decrease to 1/2 of normal value. Six representative time points from T0 to T600 show the initial state and subsequent T cell cluster expansion (also shown in Supplementary Video 7). The results show that the T cell clones are more likely to expand and generate white patches on the normal chemotactic region. Both white patches and T cell clone patterns are highly correlated with the regional fibroblast variants. e, f, QPCR analysis validated high knockdown efficiency (e) and the effect blocking IFNγ downstream signal (f) in vitro for shRNAs targeting IFNGR1, JAK1, or STAT1. g, Representative epidermis whole-mount immunofluorescent staining images and density plot of shRNA-mediated knockdown assay, relative to the corresponding dermis in Fig. 4k. h, Box-whisker plots and correlation analysis of T cell number versus percentage of infected fibroblasts (upper panels), and melanocyte number versus percentage of infected fibroblasts (lower panels) in each scale of in vivo mosaic fibroblast knockdown experiment. i, Scatter plots of median of melanocyte number versus percentage of infected fibroblasts in each scale of in vivo mosaic fibroblast knockdown experiment. Scale bars, 500 µm(g). For exact p values, see Source Data. For statistics, p summary and sample sizes, see Methods.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1–8 and their accompanying legends.
41586_2021_4221_MOESM3_ESM.mp4
Supplementary Video 1 Modelling with normal chemotaxis. The mathematical model predicts one T cell clone expansion under normal chemotaxis.
41586_2021_4221_MOESM4_ESM.mp4
Supplementary Video 2 Modelling with normal chemotaxis in large scale. The mathematical model predicts large-scale T cell clone expansion under normal chemotaxis.
41586_2021_4221_MOESM5_ESM.mp4
Supplementary Video 3 Modelling with no chemotaxis. The mathematical model predicts one T cell clone expansion under no chemotaxis.
41586_2021_4221_MOESM6_ESM.mp4
Supplementary Video 4 Modelling with no chemotaxis in larger scale. The mathematical model predicts large-scale T cell clone expansion under no chemotaxis.
41586_2021_4221_MOESM7_ESM.mp4
Supplementary Video 5 Modelling with weak chemotaxis. The mathematical model predicts one T cell clone expansion when chemotaxis decreases to half of the normal value.
41586_2021_4221_MOESM8_ESM.mp4
Supplementary Video 6 Modelling with weak chemotaxis in larger scale. The mathematical model predicts large-scale T cell clone expansion when chemotaxis decreases to half of the normal value.
41586_2021_4221_MOESM9_ESM.mp4
Supplementary Video 7 Modelling with regional variant chemotaxis. The mathematical model predicts the T cell clone distribution pattern under regional variant fibroblasts with different levels of chemotactic effect to T cell. In this model, the fibroblasts marked in blue have normal chemotactic effect, whereas those marked in green have weak chemotactic effect. In this region, T cell chemotaxis decreases to half of the normal value.
Source data
Rights and permissions
About this article
Cite this article
Xu, Z., Chen, D., Hu, Y. et al. Anatomically distinct fibroblast subsets determine skin autoimmune patterns. Nature 601, 118–124 (2022). https://doi.org/10.1038/s41586-021-04221-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-04221-8
This article is cited by
-
Identification and validation of RNA-binding protein SLC3A2 regulates melanocyte ferroptosis in vitiligo by integrated analysis of single-cell and bulk RNA-sequencing
BMC Genomics (2024)
-
Local and systemic mechanisms that control the hair follicle stem cell niche
Nature Reviews Molecular Cell Biology (2024)
-
Single-cell sequencing reveals increased LAMB3-positive basal keratinocytes and ZNF90-positive fibroblasts in autologous cultured epithelium
Communications Biology (2024)
-
Pressure and Skin: A Review of Disease Entities Driven or Influenced by Mechanical Pressure
American Journal of Clinical Dermatology (2024)
-
MYC promotes fibroblast osteogenesis by regulating ALP and BMP2 to participate in ectopic ossification of ankylosing spondylitis
Arthritis Research & Therapy (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.