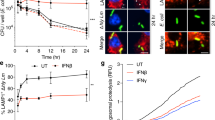
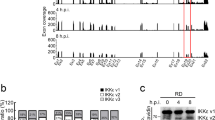
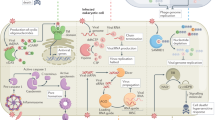
Abstract
Pathogens use virulence factors to inhibit the immune system1. The guard hypothesis2,3 postulates that hosts monitor (or ‘guard’) critical innate immune pathways such that their disruption by virulence factors provokes a secondary immune response1. Here we describe a ‘self-guarded’ immune pathway in human monocytes, in which guarding and guarded functions are combined in one protein. We find that this pathway is triggered by ICP0, a key virulence factor of herpes simplex virus type 1, resulting in robust induction of anti-viral type I interferon (IFN). Notably, induction of IFN by ICP0 is independent of canonical immune pathways and the IRF3 and IRF7 transcription factors. A CRISPR screen identified the ICP0 target MORC34 as an essential negative regulator of IFN. Loss of MORC3 recapitulates the IRF3- and IRF7-independent IFN response induced by ICP0. Mechanistically, ICP0 degrades MORC3, which leads to de-repression of a MORC3-regulated DNA element (MRE) adjacent to the IFNB1 locus. The MRE is required in cis for IFNB1 induction by the MORC3 pathway, but is not required for canonical IFN-inducing pathways. As well as repressing the MRE to regulate IFNB1, MORC3 is also a direct restriction factor of HSV-15. Our results thus suggest a model in which the primary anti-viral function of MORC3 is self-guarded by its secondary IFN-repressing function—thus, a virus that degrades MORC3 to avoid its primary anti-viral function will unleash the secondary anti-viral IFN response.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
RNA-seq and ATAC-seq data that support the findings of the study have been deposited in the Gene Expression Omnibus under accession GSE183011. CRISPR–Cas9 screen data have been deposited in the Sequence Read Archive under accession PRJNA759267. Source data are provided with this paper.
Change history
11 March 2022
In the version of this article initially published, in Extended Data Fig. 1b, the colour bars and x-axis baseline in the histogram were vertically dissociated from the data points. The colour bars and baseline have been realigned for clarity.
References
Lopes Fischer, N., Naseer, N., Shin, S. & Brodsky, I. E. Effector-triggered immunity and pathogen sensing in metazoans. Nat. Microbiol. 5, 14–26 (2020).
Jones, J. D. & Dangl, J. L. The plant immune system. Nature 444, 323–329 (2006).
Van der Biezen, E. A. & Jones, J. D. Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem. Sci. 23, 454–456 (1998).
Sloan, E. et al. Analysis of the SUMO2 proteome during HSV-1 infection. PLoS Pathog. 11, e1005059 (2015).
Sloan, E., Orr, A. & Everett, R. D. MORC3, a component of PML nuclear bodies, has a role in restricting herpes simplex virus 1 and human cytomegalovirus. J. Virol. 90, 8621–8633, https://doi.org/10.1128/JVI.00621-16 (2016).
Janeway, C. A., Jr Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 54, 1–13 (1989).
Hopfner, K. P. & Hornung, V. Molecular mechanisms and cellular functions of cGAS–STING signalling. Nat. Rev. Mol. Cell Biol. 21, 501–521 (2020).
Ishikawa, H., Ma, Z. & Barber, G. N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461, 788–792 (2009).
Scherer, M. & Stamminger, T. Emerging role of PML nuclear bodies in innate immune signaling. J. Virol. 90, 5850–5854 (2016).
Everett, R. D., Boutell, C. & Hale, B. G. Interplay between viruses and host sumoylation pathways. Nat. Rev. Microbiol. 11, 400–411 (2013).
Sacks, W. R. & Schaffer, P. A. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J. Virol. 61, 829–839 (1987).
Rapino, F. et al. C/EBPα induces highly efficient macrophage transdifferentiation of B lymphoma and leukemia cell lines and impairs their tumorigenicity. Cell Rep. 3, 1153–1163 (2013).
Ullman, A. J. & Hearing, P. Cellular proteins PML and Daxx mediate an innate antiviral defense antagonized by the adenovirus E4 ORF3 protein. J. Virol. 82, 7325–7335 (2008).
Everett, R. D. Construction and characterization of herpes simplex virus type 1 mutants with defined lesions in immediate early gene 1. J. Gen. Virol. 70, 1185–1202 (1989).
Lopez, P., Van Sant, C. & Roizman, B. Requirements for the nuclear-cytoplasmic translocation of infected-cell protein 0 of herpes simplex virus 1. J. Virol. 75, 3832–3840 (2001).
Crowl, J. T. & Stetson, D. B. SUMO2 and SUMO3 redundantly prevent a noncanonical type I interferon response. Proc. Natl Acad. Sci. USA 115, 6798–6803 (2018).
Decque, A. et al. Sumoylation coordinates the repression of inflammatory and anti-viral gene-expression programs during innate sensing. Nat. Immunol. 17, 140–149 (2016).
Ferri, F. et al. TRIM33 switches off Ifnb1 gene transcription during the late phase of macrophage activation. Nat. Commun. 6, 8900 (2015).
Malakhova, O. A. et al. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 25, 2358–2367 (2006).
Jadhav, G., Teguh, D., Kenny, J., Tickner, J. & Xu, J. Morc3 mutant mice exhibit reduced cortical area and thickness, accompanied by altered haematopoietic stem cells niche and bone cell differentiation. Sci. Rep. 6, 25964 (2016).
Li, D. Q., Nair, S. S. & Kumar, R. The MORC family: new epigenetic regulators of transcription and DNA damage response. Epigenetics 8, 685–693 (2013).
Rosendorff, A. et al. NXP-2 association with SUMO-2 depends on lysines required for transcriptional repression. Proc. Natl Acad. Sci. USA 103, 5308–5313 (2006).
Mimura, Y., Takahashi, K., Kawata, K., Akazawa, T. & Inoue, N. Two-step colocalization of MORC3 with PML nuclear bodies. J. Cell Sci. 123, 2014–2024 (2010).
Ver, L. S., Marcos-Villar, L., Landeras-Bueno, S., Nieto, A. & Ortin, J. The cellular factor NXP2/MORC3 is a positive regulator of influenza virus multiplication. J. Virol. 89, 10023–10030 (2015).
Zhang, Y. et al. MORC3 is a target of the influenza A viral protein NS1. Structure 27, 1029–1033.e1023 (2019).
Groh, S. et al. Morc3 silences endogenous retroviruses by enabling Daxx-mediated H3.3 incorporation. Nat. Commun. 12, 5996 (2021).
Yu, P. et al. Nucleic acid-sensing Toll-like receptors are essential for the control of endogenous retrovirus viremia and ERV-induced tumors. Immunity 37, 867–879 (2012).
Lopez-Flores, I. & Garrido-Ramos, M. A. The repetitive DNA content of eukaryotic genomes. Genome Dyn. 7, 1–28 (2012).
Gaidt, M. M. et al. Human monocytes engage an alternative inflammasome pathway. Immunity 44, 833–846 (2016).
Gaidt, M. M. et al. The DNA inflammasome in human myeloid cells is initiated by a STING-cell death program upstream of NLRP3. Cell 171, 1110–1124.e1118 (2017).
Blaho, J. A., Morton, E. R. & Yedowitz, J. C. Herpes simplex virus: propagation, quantification, and storage. Curr. Protoc. Microbiol. Ch. 14, Unit 14E.11 (2005).
Schmidt, T., Schmid-Burgk, J. L., Ebert, T. S., Gaidt, M. M. & Hornung, V. Designer nuclease-mediated generation of knockout THP1 cells. Methods Mol. Biol. 1338, 261–272 (2016).
Schmid-Burgk, J. L. et al. OutKnocker: a web tool for rapid and simple genotyping of designer nuclease edited cell lines. Genome Res. 24, 1719–1723 (2014).
Labun, K. et al. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 47, W171–W174 (2019).
Schmidt, T., Schmid-Burgk, J. L. & Hornung, V. Synthesis of an arrayed sgRNA library targeting the human genome. Sci. Rep. 5, 14987 (2015).
Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014).
Bartok, E. et al. iGLuc: a luciferase-based inflammasome and protease activity reporter. Nat. Methods 10, 147–154 (2013).
Spahn, P. N. et al. PinAPL-Py: a comprehensive web-application for the analysis of CRISPR/Cas9 screens. Sci. Rep. 7, 15854 (2017).
Andrews, S. FastQC: a quality control tool for high throughput sequence data. (2010).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinf. 12, 323 (2011).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Liberzon, A. et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011).
Buenrostro, J. D., Wu, B., Chang, H. Y. & Greenleaf, W. J. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21–29 (2015).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Amemiya, H. M., Kundaje, A. & Boyle, A. P. The ENCODE blacklist: identification of problematic regions of the genome. Sci. Rep. 9, 9354 (2019).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218, https://doi.org/10.1038/nmeth.2688 (2013).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Cavalcante, R. G. & Sartor, M. A. annotatr: genomic regions in context. Bioinformatics 33, 2381–2383 (2017).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Criscione, S. W., Zhang, Y., Thompson, W., Sedivy, J. M. & Neretti, N. Transcriptional landscape of repetitive elements in normal and cancer human cells. BMC Genomics 15, 583 (2014).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Freund, E. C. et al. Efficient gene knockout in primary human and murine myeloid cells by non-viral delivery of CRISPR–Cas9. J. Exp. Med. 217, e20191692 (2020).
Acknowledgements
We thank members of the Vance and Barton laboratories for advice and discussions; L. Coscoy, B. Glaunsinger and G. Barton for comments on the manuscript; D. Stetson, T. Graf, R. Tjian, X. Darzacq, D. Raulet and V. Hornung for cell lines; D. Kotov for the plasmid pMCP; B. Roizman for HSV-1 BACs; P. Dietzen and R. Chavez for technical assistance; A. Valeros and H. Nolla for assistance with automated cell sorting; M. West and P. He for assistance with robotics-based liquid handling; and F. Ramirez, C. Rose, C. Hann-Soden and S. McDevitt for deep sequencing. M.M.G. was supported by an EMBO Postdoctoral Fellowship (ALTF 1082-2018). R.E.V. is an Investigator of the Howard Hughes Medical Institute and is funded by NIH grants AI075039, AI063302 and AI155634.
Author information
Authors and Affiliations
Contributions
Conceptualization: M.M.G. and R.E.V. Investigation: M.M.G. Data analysis: M.M.G. and A.M. Writing: M.M.G. and R.E.V., with input from all co-authors. Resources: M.F. and J.K. Funding acquisition: R.E.V. and N.Y. Supervision: N.Y. and R.E.V.
Corresponding authors
Ethics declarations
Competing interests
R.E.V. consults for Ventus Therapeutics and Tempest Therapeutics. A provisional patent application covering these findings has been submitted.
Additional information
Peer review information Nature thanks John Schoggins and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Redundancy between DNA- and ICP0-triggered sensing of HSV-1 in BLaER1 monocytes.
(a-c) BLaER1 monocytes were infected with HSV-1 at MOI = 1 for 24h or transfected with DNA for 3h. Gene-expression analysis is depicted as mean + SEM of three independent experiments. (d) Immunoblot of BLaER1 monocytes of indicated genotypes. (e) BLaER1 monocytes were infected with HSV-1 at MOI = 1 for 24h and IFNβ secretion was measured by ELISA. Data is depicted as mean + SEM of four independent experiments. (f) BLaER1 monocytes were infected with HSV-1 at MOI = 5. Gene-expression is depicted as mean + SEM of three independent experiments. * p < 0.05; ** p < 0.01; *** p < 0.001; ns = not significantly different than WT, tested by two-way ANOVA and Dunnett’s or Bonferroni’s post hoc test. See Source Data for exact p values
Extended Data Fig. 2 Redundancy between DNA- and ICP0-triggered sensing of HSV-1 in THP1 and U937 cells.
PMA-differentiated THP1 (a, c) or U937 (b) human myeloid-like cells were infected with HSV-1 at MOI = 1 or indicated MOI for 24h (a, b) or indicated timepoints (c) or transfected with 2ʹ−3ʹ-cGAMP for 4h. Gene-expression analysis is depicted as mean + SEM of three independent experiments. ND = not detected. * p < 0.05; ** p < 0.01; *** p < 0.001; ns = not significantly different than WT, tested by two-way ANOVA or Mixed-effects model and Bonferroni’s post hoc test. See Source Data for exact p values
Extended Data Fig. 3 HSV-1 ΔICP0 is attenuated in BLaER1 monocytes.
(a) BLaER1 monocytes were infected with HSV-1 at indicated MOI and protein expression was analyzed by immunoblot at indicated timepoints. Data is depicted as one representative of two experiments. (b) BLaER1 monocytes were infected with HSV-1 at MOI = 1 and viral progeny was quantified in the supernatant. Mean of five independent experiments is shown. (c) BLaER1 monocytes were infected with HSV-1 at indicated MOI and analyzed by flow cytometry. Mean + SEM of three independent experiments is depicted. Data is duplicated in Fig. 3b. * p < 0.05; *** p < 0.001; ns = not significantly different than HSV-1 WT, tested by two-way ANOVA and Bonferroni’s post hoc test. See Source Data for exact p values
Extended Data Fig. 4 Virulence factors induce IFN in monocytes independently of PRR-signaling hubs.
BLaER1 monocytes (a), PMA-differentiated THP1 (b) or U937 (c) cells expressing doxycycline-inducible virulence factors were stimulated with doxycycline for 24h, left untreated or stimulated with DNA for 3h. Gene-expression as quantified by q-RT-PCR is depicted as mean + SEM of three independent experiments. IFNB1 expression levels in (a) are partially duplicated from Fig. 1c. * p < 0.05; ** p < 0.01; *** p < 0.001; ns = not significantly different than the corresponding mCherry-expressing condition, tested by two-way ANOVA or Mixed-effects model and Dunnett’s post hoc test. ND = not detected. See Source Data for exact p values
Extended Data Fig. 5 Virulence factor activity is required for IFN induction.
(a, b) BLaER1 monocytes expressing doxycycline-inducible virulence factors were stimulated with doxycycline for 24h. Gene-expression as quantified by q-RT-PCR is depicted as mean ± SEM of three independent experiments and protein expression by immunoblot is depicted from one representative experiment of two. *** p < 0.001; ns = not significantly different than the corresponding mCherry-expressing condition, tested by two-way ANOVA or Mixed-effects model and Dunnett’s post hoc test. See Source Data for exact p values. (c-f) BLaER1 monocytes expressing doxycycline-inducible virulence factors were stimulated with doxycycline for 24h. Transcriptomic changes in BLaER1 monocytes as detected by RNA-seq were analyzed by PCA of variance stabilizing transformed counts. Top 10 genes that contribute to individual PC directions are depicted (c). Heatmap of log normalized counts of the top 40 most variable genes (column normalized) from three independent RNA-seq experiments is shown (e). Modules from the Molecular Signatures Database that were found to be enriched in differentially expressed genes upon virulence factor expression in BLaER1 monocytes (f). Genes contributing to individual modules can be found in Supplementary Table 1
Extended Data Fig. 6 Validation of MORC3 as a repressor of IFN.
(a) Schematic of a genome wide CRISPR screen to identify negative regulators of IFN. (b) PinAPL.py analysis of the CRISPR screen to identify negative regulators of IFN. Significantly enriched genes are labeled. Raw data can be found in Supplementary Table 2. (c) U2OS cells were infected with HSV-1 for 24h. One representative immunoblot of two is shown. (d, e) Gene expression of BLaER1 monocytes is shown as mean + SEM of 3 independent experiment from one representative clone or two (multiple KOs) or one clone (WT and MORC3–/–) except CXCL10 quantification in (d) which is shown as mean of 2 independent experiments. *** p < 0.001; significantly different than the corresponding WT condition, tested by two-way ANOVA and Dunnett’s post hoc test. See Source Data for exact p values. (f-i) Immunoblot of BLaER1 monocytes of indicated genotypes
Extended Data Fig. 7 Characterization of MORC3 deficiency in THP1, U937 and BLaER1 cells.
(a-d) Gene-expression in PMA-differentiated THP1-Cas9 or PMA-differentiated U937-Cas9 human myeloid-like cells expressing indicated sgRNAs is depicted as mean + SEM of three independent experiments. Protein expression in the same cells was analyzed by immunoblot. ND = not detected. *** p < 0.001; ns = not significantly different than the corresponding scramble sgRNA-expressing condition, tested by two-way ANOVA and Bonferroni’s post hoc test. See Source Data for exact p values. (e) Transcriptional changes in BLaER1 monocytes as detected by RNA-seq in three independent experiments are depicted by PCA. These data are partially duplicated from Extended Data Fig. 5d. (f) Modules from the Molecular Signatures Database that were found to be enriched in differentially expressed genes in MORC3–/– BLaER1 monocytes. Background gene set includes all genes that have base mean expression of at least 1 in MORC3–/– BLaER1 monocytes. Genes contributing to individual modules can be found in Supplementary Table 1
Extended Data Fig. 8 PML is not required for IFNB1 regulation by MORC3.
(a) Gene expression of BLaER1 monocytes of indicated genotype is depicted as mean + SEM of three independent experiments. (b) Protein expression of BLaER1 monocytes was analyzed by immunoblot. (c) BLaER1 monocytes expressing doxycycline-inducible virulence factors were stimulated with doxycycline for 24h. Gene-expression as quantified by q-RT-PCR is depicted as mean + SEM of three independent experiments. (d, e) Gene-expression in HCT116-Cas9 cells expressing indicated sgRNAs is depicted as mean + SEM of three independent experiments. Protein expression in the same cells was analyzed by immunoblot. ND = not detected. ns = not significantly different than the corresponding scramble sgRNA-expressing condition, tested by Mixed-effects model and Bonferroni’s post hoc test. See Source Data for exact p values. (f) BLaER1 monocytes of indicated genotype were infected with ΔICP0 HSV-1 at indicated MOI for 6h and analyzed by flow cytometry. Mean + SEM of three independent experiment is depicted. * p < 0.05 ; ns = not significantly different than the IFNAR1–/–IFNAR2–/– condition, tested by two-way ANOVA and Bonferroni’s post hoc test. See Source Data for exact p values
Extended Data Fig. 9 Quantification of expression of ERV families.
ERV family expression was quantified with RepEnrich2 in IFNAR1–/–IFNAR2–/– mCherry vs MORC3–/–IFNAR1–/–IFNAR2–/– monocytes. ERVs with an FDR <0.05 are highlighted in red. The recommended RepEnrich2-EdgeR pipeline did not detect any regulation of ERVs upon MORC3 deficiency. RepEnrich2-DeSeq2 analysis suggested minimal up- and down-regulation of ERV families. The only strongly de-repressed ERV family upon MORC3 deficiency includes members within the MORC3-repressed region on chromosome 9, consistent with a positional rather than an ERV-specific de-repression
Extended Data Fig. 10 MORC3 deficiency leads to de-repression of a gene cluster at chromosome 9.
(a) Column-normalized heatmap analysis of transcriptomic changes in BLaER1 monocytes as detected by RNA-seq in three independent experiments. (b) Transcriptional changes in BLaER1 monocytes as detected by RNA-seq in three independent experiments are depicted by PCA. These data are partially duplicated from Extended Data Fig. 7e. (c) Log transcripts per million (TPM) of genes in BLaER1 monocytes of genes clustered near IFNB1 on chromosome 9 as detected by RNA-seq from three independent experiments. All protein coding genes within this region are depicted. (d) BLaER1 monocytes were stimulated with indicated PAMPs for 3h or 24h. Gene-expression analysis is depicted as mean + SEM of five independent experiments. (e) Log transcripts per million (TPM) of IFN genes in BLaER1 monocytes were detected by RNA-seq in three independent experiments. (f, g) BLaER1 monocytes were stimulated with DNA for 12h or left untreated. Cytokine secretion or gene expression from one representative clone of two per genotype except WT is shown as mean + SEM of four (f) or three (g) independent experiments. * p < 0.05 ; *** p < 0.001; ns = not significantly different than WT (unless otherwise indicated), tested by two-way ANOVA and Bonferroni’s post hoc test. See Source Data for exact p values. (h) Gene expression from BLaER1 monocytes of indicated genotypes is depicted as mean + SEM of three independent experiments. *** p < 0.001; ns = not significantly different than WT, tested by two-way ANOVA and Dunnett’s post hoc test. See Source Data for exact p values. (i, j) Transcriptomic changes in BLaER1 monocytes as detected by RNA-seq in three independent experiments are depicted by PCA. The data in (i) is partially duplicated from (b). The distance on PC1-axis between samples and the mean of mCherry-expressing cells was calculated and is depicted as mean + SEM from three independent experiments. *** p < 0.001 significantly different than WT, tested by two-way ANOVA and Dunnett’s post hoc test. See Source Data for exact p values
Extended Data Fig. 11 Positional de-repression of IFNB1 explains IFN induction by virulence factors.
(a) BLaER1 monocytes were infected with HSV-1 at MOI=1 for 24h. MLLT3 expression is depicted as mean + SEM of n = 3 independent experiments. *** p < 0.001; ns = not significantly different than mock infection, tested by two-way ANOVA and Dunnett’s post hoc test. See Source Data for exact p values. (b) Mean log transcripts per million (TPM) of a gene cluster at chromosome 9 in BLaER1 monocytes. Mean TPM is calculated for each condition across three independent experiments. All protein coding genes within this region are depicted. (c) Log transcripts per million (TPM) of IFN genes in BLaER1 monocytes upon virulence factor expression were detected by RNA-seq in three independent experiments. (d) BLaER1 monocytes expressing doxycycline-inducible virulence factors were stimulated with doxycycline for 24h. Gene expression is shown as mean + SEM of n = 3 independent experiments. ** p < 0.01; *** p < 0.001; ns = not significantly different than WT, tested by two-way ANOVA and Dunnett’s post hoc test. See Source Data for exact p values. Note that IFNB1–/– cells harbor small indels within IFNB1 that allow detection of mRNA by q-RT-PCR. (e, f) BLaER1 monocytes expressing doxycycline-inducible virulence factors were stimulated with doxycycline for 24h. Transcriptomic changes as detected by RNA-seq in three independent experiments were analyzed PCA. Data is partially duplicated from Extended Data Fig. 10i. The distance on PC1-axis between samples and the mean of mCherry-expressing cells was calculated and is depicted as mean + SEM from three independent experiments. *** p < 0.001 significantly different than WT, tested by two-way ANOVA and Dunnett’s post hoc test. See Source Data for exact p values
Extended Data Fig. 12 Validation of the MORC3-regulated DNA element.
(a, b) STAT1–/–STAT2–/–BLaER1-Cas9 expressing a randomly integrated IFNB1-promoter-Luciferase reporter were transduced with the indicated sgRNAs and stimulated with cytosolic DNA for 24h. Luciferase signal and IFNβ secretion is depicted as mean + SEM of n = 3 independent experiments. * p < 0.05; ns = not significantly different, tested by paired, two-sided t-test. See Source Data for exact p values. (c) Protein expression of indicated BLaER1 monocytes was analyzed by immunoblot. This experiment was performed once. (d) Consensus sequences of amplicon sequencing at the MRE locus from STAT1–/–STAT2–/– MREΔ/Δ BLaER1 Cas9 cells were aligned to the WT reference. 3166bp were omitted from the reference sequence. (e) Protein expression of indicated BLaER1 monocytes was analyzed by immunoblot. One representative experiment of two is depicted. (f) Gene expression of indicated BLaER1 monocytes is depicted as mean + SEM of three independent experiments from one representative clone of two (MORC3–/– MLLT3–/– IFNAR1–/– IFNAR2–/– and MORC3–/– FOCAD–/– IFNAR1–/– IFNAR2–/–) or one (IFNAR1–/–IFNAR2–/– and MORC3–/–IFNAR1–/–IFNAR2–/–) per genotype. ns = not significantly different than the IFNAR1–/–IFNAR2–/– condition, tested by two-way ANOVA and Dunnett’s post hoc test. See Source Data for exact p values. (g) Protein expression of indicated BLaER1 monocytes was analyzed by immunoblot. This experiment was performed once. (h) Log transcripts per million (TPM) of high-confidence MORC3 targets (genes de-repressed in MORC3–/– IFNAR1–/–IFNAR2–/–, MORC3–/– IFNB1–/– and MORC3–/– STAT1–/–STAT2–/– Cas9) in WT = MORC3–/– STAT1–/–STAT2–/– Cas9 or indicated genotypes are depicted as mean of two independent RNA-seq experiments. (i) STAT1–/–STAT2–/– Cas9 (“WT”) or STAT1–/–STAT2–/– MREΔ/Δ BLaER1 Cas9 were infected with HSV-1 at indicated MOI for 4-5h. One representative immunoblot of two is depicted. (j) Peripheral blood human monocytes were nucleofected with indicated sgRNA:Cas9 complexes and differentiated with M-CSF for 5 days. Gene expression from three independent donors is depicted as mean + SEM. * p < 0.05; ** p < 0.01; *** p < 0.001; significantly different than the scramble sgRNA condition, tested by two-way ANOVA and Bonferroni’s post hoc test. ND = not detected. Source Data for exact p values
Supplementary information
Supplementary Information
This file contains Supplementary Figs. 1–4 (showing FACS gating strategies and the full scans of the immunoblots), Supplementary Note 1 and Supplementary References.
Supplementary Table 1
Gene module enrichment in RNA-seq data.
Supplementary Table 2
Candidate negative regulators of IFN. PinAPL.py analysis of the CRISPR screen.
Source data
Rights and permissions
About this article
Cite this article
Gaidt, M.M., Morrow, A., Fairgrieve, M.R. et al. Self-guarding of MORC3 enables virulence factor-triggered immunity. Nature 600, 138–142 (2021). https://doi.org/10.1038/s41586-021-04054-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-04054-5
This article is cited by
-
Conservation and similarity of bacterial and eukaryotic innate immunity
Nature Reviews Microbiology (2024)
-
Sensing microbial infections in the Drosophila melanogaster genetic model organism
Immunogenetics (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.