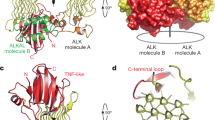
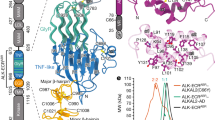
Abstract
Anaplastic lymphoma kinase (ALK)1 and the related leukocyte tyrosine kinase (LTK)2 are recently deorphanized receptor tyrosine kinases3. Together with their activating cytokines, ALKAL1 and ALKAL24,5,6 (also called FAM150A and FAM150B or AUGβ and AUGα, respectively), they are involved in neural development7, cancer7,8,9 and autoimmune diseases10. Furthermore, mammalian ALK recently emerged as a key regulator of energy expenditure and weight gain11, consistent with a metabolic role for Drosophila ALK12. Despite such functional pleiotropy and growing therapeutic relevance13,14, structural insights into ALK and LTK and their complexes with cognate cytokines have remained scarce. Here we show that the cytokine-binding segments of human ALK and LTK comprise a novel architectural chimera of a permuted TNF-like module that braces a glycine-rich subdomain featuring a hexagonal lattice of long polyglycine type II helices. The cognate cytokines ALKAL1 and ALKAL2 are monomeric three-helix bundles, yet their binding to ALK and LTK elicits similar dimeric assemblies with two-fold symmetry, that tent a single cytokine molecule proximal to the cell membrane. We show that the membrane-proximal EGF-like domain dictates the apparent cytokine preference of ALK. Assisted by these diverse structure–function findings, we propose a structural and mechanistic blueprint for complexes of ALK family receptors, and thereby extend the repertoire of ligand-mediated dimerization mechanisms adopted by receptor tyrosine kinases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Coordinates and structure factors for the complexes have been deposited in the Protein Data Bank (PDB) under accessions 7NWZ (ALKTG–ALKAL2), 7NX0 (LTKTG–ALKAL1–Nb3.16), 7NX1 (LTKTG), 7NX2 (unbound Fab324), 7NX3 (ALKTG–EGFL–Fab324), 7NX4 (ALKTG–EGFL). Most common single-nucleotide polymorphisms were obtained from the COSMIC database (https://cancer.sanger.ac.uk/cosmic) and gnomAD database (https://gnomad.broadinstitute.org). The Ba/F3 cell lines expressing ALK(WT), ALK(R753Q) or ALK(F856S) can be obtained from the authors upon request. Source data are provided with this paper.
References
Morris, S. W. et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science 263, 1281–1284 (1994).
Ben-Neriah, Y. & Bauskin, A. R. Leukocytes express a novel gene encoding a putative transmembrane protein-kinase devoid of an extracellular domain. Nature 333, 672–676 (1988).
Zhang, H. et al. Deorphanization of the human leukocyte tyrosine kinase (LTK) receptor by a signaling screen of the extracellular proteome. Proc. Natl Acad. Sci. USA 111, 15741–15745 (2014).
Reshetnyak, A. V. et al. Augmentor α and β (FAM150) are ligands of the receptor tyrosine kinases ALK and LTK: Hierarchy and specificity of ligand-receptor interactions. Proc. Natl Acad. Sci. USA 112, 15862–15867 (2015).
Guan, J. et al. FAM150A and FAM150B are activating ligands for anaplastic lymphoma kinase. eLife 4, e09811 (2015).
Reshetnyak, A. V. et al. Identification of a biologically active fragment of ALK and LTK-ligand 2 (augmentor-α). Proc. Natl Acad. Sci. USA 115, 8340–8345 (2018).
Hallberg, B. & Palmer, R. H. Mechanistic insight into ALK receptor tyrosine kinase in human cancer biology. Nat. Rev. Cancer 13, 685–700 (2013).
Janostiak, R., Malvi, P. & Wajapeyee, N. Anaplastic lymphoma kinase confers resistance to BRAF kinase inhibitors in melanoma. iScience 16, 453–467 (2019).
Javanmardi, N. et al. Analysis of ALK, MYCN, and the ALK ligand ALKAL2 (FAM150B/AUGα) in neuroblastoma patient samples with chromosome arm 2p rearrangements. Genes Chromosomes Cancer 59, 50–57 (2020).
Li, N. et al. Gain-of-function polymorphism in mouse and human Ltk: implications for the pathogenesis of systemic lupus erythematosus. Hum. Mol. Genet. 13, 171–179 (2004).
Orthofer, M. et al. Identification of ALK in thinness. Cell 181, 1246–1262.e22 (2020).
Pospisilik, J. A. et al. Drosophila genome-wide obesity screen reveals Hedgehog as a determinant of brown versus white adipose cell fate. Cell 140, 148–160 (2010).
Borenäs, M. et al. ALK ligand ALKAL2 potentiates MYCN‐driven neuroblastoma in the absence of ALK mutation. EMBO J. 40, e105784 (2021).
Sano, R. et al. An antibody-drug conjugate directed to the ALK receptor demonstrates efficacy in preclinical models of neuroblastoma. Sci. Transl. Med. 11, eaau9732 (2019).
Dornburg, A. et al. Comparative genomics within and across bilaterians illuminates the evolutionary history of ALK and LTK proto-oncogene origination and diversification. Genome Biol. Evol. 13, evaa228 (2021).
Murray, P. B. et al. Heparin is an activating ligand of the orphan receptor tyrosine kinase ALK. Sci. Signal. 8, ra6 (2015).
Alvarado, D. et al. Anti-ALK antibodies and methods for use thereof. US patent 15/755421 (2021).
Crick, F. H. C. & Rich, A. Structure of polyglycine II. Nature 176, 780–781 (1955).
Lorén, C. E. et al. A crucial role for the anaplastic lymphoma kinase receptor tyrosine kinase in gut development in Drosophila melanogaster. EMBO Rep. 4, 781–786 (2003).
Kolodny, R. Searching protein space for ancient sub-domain segments. Curr. Opin. Struct. Biol. 68, 105–112 (2021).
Fadeev, A. et al. ALKALs are in vivo ligands for ALK family receptor tyrosine kinases in the neural crest and derived cells. Proc. Natl Acad. Sci. USA 115, E630–E638 (2018).
Wang, Y. W. et al. Identification of oncogenic point mutations and hyperphosphorylation of anaplastic lymphoma kinase in lung cancer. Neoplasia 13, 704–715 (2011).
Maxson, J. E. et al. Therapeutically targetable ALK mutations in leukemia. Cancer Res. 75, 2146–2150 (2015).
Durham, B. H. et al. Activating mutations in CSF1R and additional receptor tyrosine kinases in histiocytic neoplasms. Nat. Med. 25, 1839–1842 (2019).
Malinauskas, T., Aricescu, A. R., Lu, W., Siebold, C. & Jones, E. Y. Modular mechanism of Wnt signaling inhibition by Wnt inhibitory factor 1. Nat. Struct. Mol. Biol. 18, 886–893 (2011).
Wehrman, T. et al. Structural and mechanistic insights into nerve growth factor interactions with the TrkA and p75 receptors. Neuron 53, 25–38 (2007).
Ogiso, H. et al. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell 110, 775–787 (2002).
Elegheert, J. et al. Extracellular complexes of the hematopoietic human and mouse CSF-1 receptor are driven by common assembly principles. Structure 19, 1762–1772 (2011).
Schlessinger, J. et al. Crystal structure of a ternary FGF–FGFR–heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol. Cell 6, 743–750 (2000).
Uchikawa, E., Choi, E., Shang, G., Yu, H. & Xiao-Chen, B. Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor–ligand complex. eLife 8, e48630 (2019).
Aricescu, A. R., Lu, W. & Jones, E. Y. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D 62, 1243–1250 (2006).
Reeves, P. J., Callewaert, N., Contreras, R. & Khorana, H. G. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc. Natl Acad. Sci. USA 99, 13419–13424 (2002).
Backliwal, G. et al. Valproic acid: A viable alternative to sodium butyrate for enhancing protein expression in mammalian cell cultures. Biotechnol. Bioeng. 101, 182–189 (2008).
Gorrec, F. The MORPHEUS II protein crystallization screen. Acta Crystallogr. F 71, 831–837 (2015).
Kabsch, W. XDS. Acta Crystallogr. D. 66, 125–132 (2010).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Bricogne G., et al. BUSTER 2.11.2 (United Kingdom Global Phasing Ltd, 2017).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Terwilliger, T. C. Maximum-likelihood density modification. Acta Crystallogr. D 56, 965–972 (2000).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. Sect. D 75, 861–877 (2019).
Langer, G., Cohen, S. X., Lamzin, V. S. & Perrakis, A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 (2008).
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D 67, 355–367 (2011).
D’Arcy, A., Villard, F. & Marsh, M. An automated microseed matrix-screening method for protein crystallization. Acta Crystallogr. D 63, 550–554 (2007).
Strong, M. et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA 103, 8060–8065 (2006).
Howarth, M. & Ting, A. Y. Imaging proteins in live mammalian cells with biotin ligase and monovalent streptavidin. Nat. Protoc. 3, 534–545 (2008).
Hopkins, J. B., Gillilan, R. E. & Skou, S. BioXTAS RAW: Improvements to a free open-source program for small-angle X-ray scattering data reduction and analysis. J. Appl. Crystallogr. 50, 1545–1553 (2017).
Biasini, M. et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252 (2014).
Webb, B. & Sali, A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinforma. 2016, 5.6.1–5.6.37 (2016).
Yoshimi, A. et al. Coordinated alterations in RNA splicing and epigenetic regulation drive leukaemogenesis. Nature 574, 273–277 (2019).
Ishihara, T. et al. HEN-1, a secretory protein with an LDL receptor motif, regulates sensory integration and learning in Caenorhabditis elegans. Cell 109, 639–649 (2002).
Englund, C. et al. Jeb signals through the Alk receptor tyrosine kinase to drive visceral muscle fusion. Nature 425, 512–516 (2003).
Lee, H. H., Norris, A., Weiss, J. B. & Frasch, M. Jelly belly protein activates the receptor tyrosine kinase Alk to specify visceral muscle pioneers. Nature 425, 507–512 (2003).
Blum, M. et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 49, D344–D354 (2021).
Pentelute, B. L. et al. X-ray structure of snow flea antifreeze protein determined by racemic crystallization of synthetic protein enantiomers. J. Am. Chem. Soc. 130, 9695–9701 (2008).
Buglino, J., Shen, V., Hakimian, P. & Lima, C. D. Structural and biochemical analysis of the Obg GTP binding protein. Structure 10, 1581–1592 (2002).
Weidenweber, S. et al. Structure of the acetophenone carboxylase core complex: Prototype of a new class of ATP-dependent carboxylases/hydrolases. Sci. Rep. 7, 1–10 (2017).
Dunne, M. et al. Salmonella phage S16 tail fiber adhesin features a rare polyglycine rich domain for host recognition. Structure 26, 1573–1582.e4 (2018).
Krissinel, E. & Henrick, K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr., Sect. D: Biol. Crystallogr. 60, 2256–2268 (2004).
Acknowledgements
We thank the staff of beamlines P13 and P14 (Petra III, Deutsches Elektronen-Synchrotron), Proxima 2A (SOLEIL) and ID23-2 (ESRF) and SWING (SOLEIL) for their technical support and beamtime allocation. S.D.M. was supported by a predoctoral fellowship from the Flanders Agency for Innovation and Entrepreneurship (VLAIO-Flanders, Belgium). Y.B. is a post-doctoral research fellow of Research Foundation Flanders (FWO). A.Y. acknowledges support from the Japan Society for the Promotion of Science (JSPS) Home-Returning Researcher Development Research (grant number 19K24691), KAKENHI (grant number 21H04828) and National Cancer Center Research and Development Funds (2020-A-2). S.N.S. acknowledges research support from the FWO (grant number G0B4918N), Ghent University (grant number BOF17-GOA-028), the Hercules Foundation (no. AUGE-11-029) and the Flanders Institute for Biotechnology (VIB).
Author information
Authors and Affiliations
Contributions
S.D.M. performed protein expression and purification and performed crystallization experiments with contributions from M.P. S.D.M. determined and analysed crystal structures with contributions from Y.B. and S.N.S. J.F. performed analysis of SAXS data with contributions from S.D.M. S.D.M. performed ITC, SEC-MALLS and BLI experiments with contributions from M.P. . O.A.-W. and A.Y. contributed materials and designed cell assays. M.K., I.O. and J.M. performed cellular assays under the supervision of A.Y. J.F.B. carried out evolutionary and structural analyses with contributions from S.D.M. and S.N.S. S.D.M. and S.N.S. wrote the manuscript with contributions from all authors. S.N.S. conceived and supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks the anonymous reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Phylogenetic analysis and evolution of ALK and LTK.
a, Phylogenetic analysis of ALK (a). Multiple sequence alignment was performed with MAFFT 7 using the E-INS-I algorithm for refinement. The phylogenetic tree was visualized using Phyl.io tool as implemented in MAFFT. ALK is an evolutionarily ancient RTK in C. elegans and D. melanogaster, where it is activated by HEN-150 and Jeb51,52. b, Sequence alignment of ALK sequences focusing on the heparin binding motif. Positively charged residues are highlighted in blue. The multiple sequence alignment was performed with MAFFT 7 using the E-INS-I algorithm for refinement. c, Phylogenetic analysis of LTK performed as for ALK. In mammals the LTK ectodomain consists of the TG and EGF-like domains while other vertebrates contain the additional MAM-LDLa-MAM domains at the N-terminus.
Extended Data Fig. 2 Purification of ALKTG-EGFL and LTKTG and structural details of the TNFL and GR subdomains of the novel TG supradomain fold.
a,b,c Representative chromatograms and SDS-PAGE gels for the purification of ALKTG-EGFL (a), ALKTG-EGFL—Fab324 (b) and LTKTG (c). The arrow indicates the shift in elution volume after EndoH digest of ALKTG-EGFL. Each protein was purified several times, chromatograms and SDS-PAGE analysis of each sample are representative for different protein batches. Uncropped gels are included in source data. d, ALKTG-EGFL structure colored according to secondary structure elements. α-Helices (blue), β-strands (green), pGII-helices (orange), loops (grey). e, Structure of the ALKTG-EGFL—Fab324 complex with ALK coloured according to its secondary structure elements. CDR loops of Fab324 are coloured yellow. The constant domains of Fab324 are omitted for clarity. f, LTKTG structure colored according to secondary structure elements. g, Hexagonal pGII-helix arrangement surrounding pGII-helix d in LTK. Vermillion pGII-helices consist exclusively of glycine residues. pGII-helix d shown as sticks, hydrogen bonds to other residues in LTK are indicated as dotted lines. The glycine-rich segment has complicated detection of a globular fold but has led to its sequence-based classification as Glycine-rich PFAM domain PF1281053. h, Schematic representations of pGII-helix arrangements in reported structures. Drastically less extensive pGII-helix arrangements than the one displayed in the GR subdomain of ALK and LTK have been observed in synthetic polyglycines20 and four functionally diverse proteins54,55,56,57. Full circles indicate pGII-helices coming out of the plane of the page while empty circles indicate helices going into the plane of the page. S16 adhesin (pdb: 6F45) Apc complex (pdb: 5L9W) obg (pdb: 5M04) Sf antifreeze protein (pdb: 3BOI). i, Sequence alignment performed with Clustal Omega of human ALK, human LTK, C. elegans ALK (SCD-2) and D. melanogaster ALK covering pGII-helices j,k and l. Residues conserved across all four species are indicated with an orange background. Conserved hydrophobic residues involved in the hydrophobic groove between the TNF-like and glycine rich region are indicated by a green sphere. j, The β-sheet subregions of ALK and a trimmed view of TNF (pdb: 1TNF) are coloured in a N-(blue) to C-terminus (red) gradient and shown side by side after structural superposition. Topology diagram for the TNFL domain of ALK and the jelly-roll fold of TNF follow the same colour scheme. Jelly-roll fold nomenclature starts with strand B according to convention. For ALKTNFL the nomenclature in black is according to the TG domain notation used in this study while the nomenclature according to the TNF convention (first β-strand labeled B) is shown in grey. Structural queries63 using the TFNL subdomains retrieved TNF/C1q-class folds (r.m.s.d =2.8 Å against C1q and TNF, 72 Cα atoms). Topology-independent searches58 covered an additional ~20 residues in the canonical TNF fold, and structure-based sequence alignments clarified the sequence homology between the A, D and E β-strands in ALK/LTKTNFL and β-strands B, E and F in TNF or TRAIL. The distinctly connected β-strands in the ALKTNFL/LTKTNFL subdomain break up the alternating sheet-to-sheet register of the TNF/C1q β-jellyroll, and instead permit the spatially contiguous sprouting of the three glycine-rich loop inserts (between β-strands D and E, F and G, and H and H’) towards the distinctive pGII-helix lattice of the ALKGR/LTKGR subdomain. The sequential B to I β-strands of the TNF/C1q β-jellyroll smoothly sew together the two β-sheets (that feature characteristic B’BIDG and FEHC faces) whereas the ALK/LTKTNFL subdomain has AIDEH” and H’HGFCB faces (primed small caps denote additional, edge β-strands). k, Annotated alignment of selected β-strands of human ALK, LTK, TNF and TRAIL. Conserved hydrophobic residues are indicated in red.
Extended Data Fig. 3 ALKAL1- and ALKAL2-dependent proliferation of Ba/F3 cells expressing ALK.
a, Molecular mass determination of ALKAL2, ALKAL1 and ALKAL1FL by size-exclusion chromatography and in-line multi-angle laser light scattering (SEC-MALLS). The differential refractive index (left vertical axis) is plotted against the determined molecular weight (right vertical axis). ALKAL2 (blue trace), ALKAL1 (green trace) and ALKAL1FL (vermillion trace). b, Cell proliferation of ALKWT or EV (empty vector) expressing Ba/F3 cells treated with 10 nM ALKAL2 with and without addition of crizotinib (n = 3 biologically independent experiments; mean ± s.d). c, Cell proliferation of Ba/F3 cells expressing ALKWT or EV upon stimulation by a concentration series of ALKAL2, ALKAL1 or ALKAL1FL at indicated concentrations. (n = 3 biologically independent experiments; mean ± s.d.; two-way ANOVA with Tukey’s multiple comparison test compared with EV; *P < 0.05, **P < 0.01 and ***P < 0.001; Exact P-values are provided in source data. The ratio of the observed ALKAL-induced cell growth with the IL-3 induced cell growth is shown in a heatmap representation for the measurements on day 4).
Extended Data Fig. 4 Biophysical characterization and purification of ALKAL-mediated complexes of ALK and LTK.
a, Calculated molecular masses for the ALK, LTK and ALKAL constructs under study. b-e, Experimental molecular mass determination of ALKAL-mediated complexes with LTKTG-EGFL (b), ALKTG-EGFL (deglycosylated) (c), LTKTG (d), ALKTG (deglycosylated) (e) by SEC-MALLS. The differential refractive index (left vertical axis) is plotted against the experimentally measured molecular mass (right vertical axis). Unbound LTK and ALK are in orange and pink respectively. Complexes with ALKAL1 or ALKAL2 are in green and blue respectively. The reported molecular mass represents the average molecular mass ± s.d. across the elution peak. f,g, Representative SEC elution profiles and SDS-PAGE analysis for the purification of the ALKTG-EGFL—ALKAL2 complex (f) and the LTKTG-EGFL—ALKAL1—Nb3.16 complex (g). Each protein was purified several times, chromatograms and SDS-PAGE analysis of each sample are representative for different protein batches. Uncropped gels are included in source data. h, Surface representation of the LTKTG-EGFL—ALKAL1—Nb3.16 complex illustrating the location of the binding sites of the non-neutralizing Nb3.16 to LTKTG far from any cytokine–receptor or receptor–receptor binding site.
Extended Data Fig. 5 Structure of ALKAL1 and ALKAL2.
a, Structural superposition of the conserved C-terminal segments of ALKAL1 and ALKAL2 (coloured in an N- (blue) to C-terminus (red) gradient) as observed in their complexes with cognate receptors. ALKAL1 and ALKAL2 share high sequence identity (66%) in their C-terminal domain, but have variable N-terminal regions. ALKAL1 residues in the interface between helix A and helices B and C are shown as sticks. αA connects via a conserved short loop to a helical hairpin constructed from αB and αC, which in turn are tethered by two conserved disulfides. A conserved stretch of ~10 residues preceding αA was not ordered in the reported structures, suggesting they might help to stabilize the soluble forms of these cytokines rather than contribute to direct receptor engagement or might help reduce the entropic cost of binding. b, Surface representation of ALKAL1 coloured according to the Eisenberg hydrophobicity scale. Hydroxyl groups surrounding the central cavity are shown as red spheres. c, Sequence alignment by ClustalOmega of human ALKAL1 with ALKAL2. Residues in the interface between helix A and helices B and C are coloured according to their position. Cysteines are coloured yellow and disulfide bonds are shown as yellow lines. d, Surface representation of the LTKTG—ALKAL1 complex with LTK in ALKAL1 coloured according to the Eisenberg hydrophobicity scale (white is more hydrophobic). The black circle denotes the ALKAL1 hydrophobic cavity. e, Multiple sequence alignments of various vertebrate ALKAL1 and ALKAL2 sequences using the ESPripT server (http://espript.ibcp.fr/ESPript/ESPript/) and structural annotation according to secondary structure elements. Symbols indicate residues participating in interaction sites 1 and 2 according to the graphical legend. Hs (Homo sapiens), Mm (Mus musculus), Gg (Gallus gallus) Xl (Xenopus laevis), Xt (Xenopus tropicalis) Dr (Danio rerio). f, ALKAL1 shown in green transparent surface and ribbon representation. Residues differing with ALKAL2 are colored vermillion and labeled with the ALKAL1/ALKAL2 numbering.
Extended Data Fig. 6 Structural details of receptor–cytokine and receptor–receptor interactions in ALK/LTK–cytokine complexes.
a, 2Fo-Fc electron density maps contoured at +1 r.m.s.d. showing details of site 1, 2 and 3 of the LTK—ALKAL1 and ALK—ALKAL2 complexes. b, Transparent surface of ALKAL1 according to the Eisenberg hydrophobicity scale illustrating similarities and differences in site 1 of LTK/ALK–cytokine complexes. Shown is the central conserved hydrophobic patch formed by leucines (L97, L116 and L120) and the interacting residues of LTK (orange). The equivalent ALK residues (pink) are shown after alignment with LTK. c, View of Site 2 in the LTK—ALKAL1 and ALK—ALKAL2 complexes. ALKALs are coloured according to the Eisenberg hydrophobicity scale. Receptor residues surrounding the hydrophobic triad of helix A (L72, F76, F80 in ALKAL1 and M93, F97, L101 in ALKAL2) are shown as sticks for LTK (orange) and ALK (pink). d, Superposition of unbound ALK (dark gray), unbound LTK (light gray), bound ALK (pink, only helices shown) and bound LTK (orange, only helices shown). e, Superposition of the ALK—ALKAL2 and LTK—ALKAL1 complexes, zoomed in on the region around the e-f loop. f, View of the site 3 groove of ALK (top) and LTK (bottom). In LTK, site 3 centers on His153, which stacks against Gly74 and Arg241 on an LTK-specific loop with Asn369 residues hydrogen bonding across the twofold axis of the complex.
Extended Data Fig. 7 Functional interrogation of site 1, site 2, and site 3 interfaces in ALK/LTK–cytokine complexes.
a,b Representative response curves as measured by biolayer-interferometry (BLI) for the interaction of wild type ALKAL1 and ALKAL1 mutants (containing charge-reversal mutations of residues involved in site 1) (a) and WT ALKAL2 and ALKAL2 mutants (b) with ALKTG-EGFL and LTK TG-EGFL. For wild type ALKALs LTK curves were fitted with a 2:1 binding model (red) while for ALK a 1:1 model was used. Start and end concentrations of the 2-fold dilution series used for the WT measurements is shown as an inset while for all mutants a 2-fold dilution series from 6.4 μM-400nM was used. c, BLI response curves for the interaction of the site 2 ALKAL1F76E mutant with LTK TG-EGFL. d, BLI response curves for the interaction of the site 2 ALKAL2F97E and ALKAL2H100A with LTKTG-EGFL. e, BLI response curves for the interaction of the site 2 ALKAL2F97E with ALKTG-EGFL. f, SDS-PAGE analysis of purified ALKAL1 and ALKAL2 mutants used in Ba/F3 and SEC-MALLS assays. Each protein was purified several times, SDS-PAGE analysis of each sample are representative for different protein batches. Uncropped gels are included in source data. g, Western blot analysis of phosphorylated ALK (Y1278 and Y1604) after stimulation with ALKAL2WT, ALKAL2R123E/R136E, ALKAL2F97E and ALKAL2H100A. Uncropped western blot scans are provided in source data. h, Capacity of ALKAL1 and ALKAL2 mutants to form complexes with LTK TG-EGFL and ALK TG-EGFL respectively as characterized by SEC-MALLS. Differential refractive index (left axis) is plotted against the determined molecular weight (right axis). LTKTG-EGFL (orange trace), LTKTG-EGFL—ALKAL1R102E/R115E (green trace) and LTKTG-EGFL—ALKAL1F76E (blue trace). ALKTG-EGFL (pink trace), ALKTG-EGFL—ALKAL2R123E/R136E (green trace) and ALKTG-EGFL—ALKAL2F97E (blue trace). The ALKAL1 site 1 mutant is unable to form a complex with ALK while the site 2 mutant still forms a binary complex. The reported molecular mass represents the average molecular mass ± s.d. across the elution peak. i, Capacity of the LTKTG-EGFLR241A LTKTG-EGFLR241A/N369G site 3 mutants to form complexes with ALKAL1 as characterized by SEC-MALLS. LTKTG-EGFLR241A (red trace), LTKTG-EGFLR241A/N369G (cyan trace), LTKTG-EGFLR241A—ALKAL1 (green trace), LTKTG-EGFLR241A/N369G—ALKAL1 (blue trace). LTKTG-EGFL (orange trace) and LTKTG-EGFL—ALKAL1 (pink trace) are shown for comparison. The reported molecular mass represents the average molecular mass ± s.d. across the elution peak. j, Cell proliferation of Ba/F3 cells expressing ALKWT or ALKM751T upon stimulation with 50 nM ALKAL1 or 50 nM ALKAL2. Western blot analysis of ALKWT or ALKM751T expression is a representative of three biologically independent experiments with similar results. Uncropped western blot scans are provided in source data.
Extended Data Fig. 8 Conservation of TG supradomains in ALK and LTK.
a, Structurally annotated multiple sequence alignments of the TG and EGFL domains of ALK and LTK using the ESPripT server (http://espript.ibcp.fr/ESPript/ESPript/). Hs (Homo sapiens), Mm (Mus musculus), Gg (Gallus gallus) Xl (Xenopus laevis), Xt (Xenopus tropicalis) Dr (Danio rerio). Symbols indicate residues participating in the different interfaces according to the graphical legend. b, Bottom view of dimerized LTK (left) and ALK (right) in surface representation coloured according to residue conservation. The ALKAL binding sites are shown as an outline of ALKAL1 (green) and ALKAL2 (blue) are shown as dashed lines. Conservation analysis was performed using the Consurf server (https://consurf.tau.ac.il) based on an alignment of 248 vertebrate sequences for ALK and 225 for LTK by COBALT. c, Top view of dimerized LTK (left) and ALK (right) coloured according to sequence conservation levels.
Extended Data Fig. 9 Mapping of missense mutations on the structures of the ALK—ALKAL2 and LTK—ALKAL1 complexes.
a, Mapping of most frequent SNPs (GnomAD) to the ALKAL2—ALKTG complex shown in top view. SNPs also found in COSMIC database (https://cancer.sanger.ac.uk/cosmic) are also indicated on the bottom view. Mutations further characterized in this study are coloured red. Inset shows the detailed position of R753 in the ALKTG—ALKAL2 complex. b, Mapping of most frequent SNPs (GnomAD) to the ALKAL1—LTKTG complex shown in top view. SNPs also found in COSMIC database (https://cancer.sanger.ac.uk/cosmic) are also indicated on the bottom view. c, Western blot analysis of the expression levels of ALKWT and the ALKR753Q and ALKF856S mutants and their ERK phosphorylation (left). On the right-side western blot analysis of phosphorylated ALKWT, ALKR753Q and ALKF856S is shown. Representative results from three biologically independent experiments with similar results. Uncropped western blot scans are provided in source data. d, Sanger sequencing of cDNA showing WT or mutant ALK expression in isogenic Ba/F3 cells. e, Cell proliferation of ALK expressing Ba/F3 cells treated with 10 nM ALKAL2 with and without addition of crizotinib for ALK R753Q and F856S mutants. Data for EV and WT ALK are repeated from Extended Data Fig. 3 for direct comparison. Crizotinib is also able to inhibit ALKAL2 induced proliferation for mutant ALK, indicating ALK dependent signalling. (n = 3 biologically independent experiments; mean ± s.d.; two-way ANOVA with Tukey’s multiple comparison test compared with DMSO control). f, Proliferation of Ba/F3 cells expressing ALK carrying the R753Q or F856S mutation upon stimulation by a concentration series of ALKAL2, ALKAL1 or ALKAL1FL at indicated concentration (n=3 biologically independent experiments; mean ± s.d.; two-way ANOVA with Tukey’s multiple comparison test compared with ALKWT. ALKAL-induced cell growth relative to that of cells cultured with IL-3 is shown in a heatmap representation. EV and ALKWT controls are included for comparison. *P < 0.05, **P < 0.01 and ***P < 0.001. Exact P-values are provided in source data).
Extended Data Fig. 10 Mechanistic insights into the assembly of ALK/LTK–cytokine complexes derived from microcalorimetry, SAXS, and SEC-MALLS.
a,b ITC experiments for the titration of LTKTG-EGFL (5 µM) with ALKAL1 (56 µM). (a) Titration of LTKTG-EGFL (12 µM) with ALKAL2 (56 µM). (b). ITC titration curves, the left panel shows the raw data with the differential electrical power (DP) plotted against time. The right panel represents the binding isotherm obtained from the integration of the raw data and fitted to a one-site model. mean ± s.d were calculated based on 3 measurements. c, Small-angle X-ray scattering analysis and calculated FoXS fits of the binary ALKTG-EGFL—ALKAL2 (pink), ternary ALKTG-EGFL—ALKAL2 (purple), binary LTKTG-EGFL—ALKAL1 (orange) and ternary LTKTG-EGFL—ALKAL1 (light orange) to experimental SAXS data (black curves). d, ITC experiments for the titration of LTKTG-EGFL (7.2 µM) with ALKAL1FL (55 µM) Data representative of 2 independent experiments. e, Ternary ALKTG:ALKAL2 complex with regions differing with ALKAL1 coloured vermillion. C-termini of the TG domains leading towards the EGF-like domains are coloured red. f, ITC experiments for the titration of LTKTG (10 µM) with ALKAL1 (70 µM) Data representative of 2 independent experiments. g, ITC experiments for the titration of LTKTG (10 µM) with ALKAL2 (100 µM) Data representative of 2 independent experiments. h, ITC experiments for the titration of LTKTG (10 µM) with ALKAL1FL (40 µM) Data representative of 2 independent experiments. i, ITC experiments for the titration of ALKFL (8 µM) with ALKAL2 (82 µM) Data representative of 2 independent experiments. j, Characterization of heparin-induced ALKFL dimerization by SEC-MALLS. Differential refractive index (left vertical axis) is plotted against the determined molecular weight (right vertical axis). ALKFL (pink), ALKFL complexes with heparin dp20 (green) and ALKFL complexed with heparin dp20 and ALKAL2 (blue trace). The reported molecular mass represents the average molecular mass ± s.d. across the elution peak. k, Overview of the different ligand-mediated extracellular assemblies across RTKs. Trka—NGF (PDB: 2IFG), EGFR—EGF (PDB: 1IVO), INSR—INS (PDB: 6PXW), CSF1R—CSF1 (PDB: 4WRM), FGFR—heparin—FGF (PDB: 1FQ9), ALKTG—ALKAL2 (PDB: 7NWZ).
Supplementary information
Supplementary Figure 1
Uncropped blots shown in Extended Data Figs. 2, 4, 7, 9.
Rights and permissions
About this article
Cite this article
De Munck, S., Provost, M., Kurikawa, M. et al. Structural basis of cytokine-mediated activation of ALK family receptors. Nature 600, 143–147 (2021). https://doi.org/10.1038/s41586-021-03959-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03959-5
This article is cited by
-
Improving de novo protein binder design with deep learning
Nature Communications (2023)
-
Placing human gene families into their evolutionary context
Human Genomics (2022)
-
Highlights of recent cancer research
Holistic Integrative Oncology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.