Abstract
Glutathione (GSH) is a small-molecule thiol that is abundant in all eukaryotes and has key roles in oxidative metabolism1. Mitochondria, as the major site of oxidative reactions, must maintain sufficient levels of GSH to perform protective and biosynthetic functions2. GSH is synthesized exclusively in the cytosol, yet the molecular machinery involved in mitochondrial GSH import remains unknown. Here, using organellar proteomics and metabolomics approaches, we identify SLC25A39, a mitochondrial membrane carrier of unknown function, as a regulator of GSH transport into mitochondria. Loss of SLC25A39 reduces mitochondrial GSH import and abundance without affecting cellular GSH levels. Cells lacking both SLC25A39 and its paralogue SLC25A40 exhibit defects in the activity and stability of proteins containing iron–sulfur clusters. We find that mitochondrial GSH import is necessary for cell proliferation in vitro and red blood cell development in mice. Heterologous expression of an engineered bifunctional bacterial GSH biosynthetic enzyme (GshF) in mitochondria enables mitochondrial GSH production and ameliorates the metabolic and proliferative defects caused by its depletion. Finally, GSH availability negatively regulates SLC25A39 protein abundance, coupling redox homeostasis to mitochondrial GSH import in mammalian cells. Our work identifies SLC25A39 as an essential and regulated component of the mitochondrial GSH-import machinery.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Proteomics data have been deposited to the ProteomeXchange Consortium with the dataset identifier PXD027673. Other data generated are available from the corresponding author upon reasonable request. Source data are provided with this paper.
References
Meister, A. & Anderson, M. E. Glutathione. Annu. Rev. Biochem. 52, 711–760 (1983).
Mårtensson, J., Lai, J. C. & Meister, A. High-affinity transport of glutathione is part of a multicomponent system essential for mitochondrial function. Proc. Natl Acad. Sci. USA 87, 7185–7189 (1990).
Deponte, M. The incomplete glutathione puzzle: just guessing at numbers and figures? Antioxid. Redox Signal. 27, 1130–1161 (2017).
Griffith, O. W. & Meister, A. Origin and turnover of mitochondrial glutathione. Proc. Natl Acad. Sci. USA 82, 4668–4672 (1985).
Hwang, C., Sinskey, A. J. & Lodish, H. F. Oxidized redox state of glutathione in the endoplasmic reticulum. Science 257, 1496–1502 (1992).
Meredith, M. J. & Reed, D. J. Status of the mitochondrial pool of glutathione in the isolated hepatocyte. J. Biol. Chem. 257, 3747–3753 (1982).
Deneke, S. M. & Fanburg, B. L. Regulation of cellular glutathione. Am. J. Physiol. Lung Cell. Mol. Physiol. 257, L163–L173 (1989).
Jones, D. P. Redox potential of GSH/GSSG couple: assay and biological significance. Methods Enzymol. 348, 93–112 (2002).
Kurosawa, K., Hayashi, N., Sato, N., Kamada, T. & Tagawa, K. Transport of glutathione across the mitochondrial membranes. Biochem. Biophys. Res. Commun. 167, 367–372 (1990).
Griffith, O. W. & Meister, A. Potent and specific inhibition of glutathione synthesis by buthionine sulfoximine (S-n-butyl homocysteine sulfoximine). J. Biol. Chem. 254, 7558–7560 (1979).
Münch, C. & Harper, J. W. Mitochondrial unfolded protein response controls matrix pre-RNA processing and translation. Nature 534, 710–713 (2016).
Lee, H.-R. et al. Adaptive response to GSH depletion and resistance to l-buthionine-(S,R)-sulfoximine: involvement of Nrf2 activation. Mol. Cell. Biochem. 318, 23–31 (2008).
Sun, X. et al. Activation of the p62–Keap1–NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology 63, 173–184 (2016).
Booty, L. M. et al. Selective disruption of mitochondrial thiol redox state in cells and in vivo. Cell Chem. Biol. 26, 449-461.e8 (2019).
Ruprecht, J. J. & Kunji, E. R. S. The SLC25 mitochondrial carrier family: structure and mechanism. Trends Biochem. Sci. 45, 244–258 (2020).
Pebay-Peyroula, E. et al. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature 426, 39–44 (2003).
Zhu, X. G. et al. CHP1 regulates compartmentalized glycerolipid synthesis by activating GPAT4. Mol. Cell 74, 45–58.e7 (2019).
Zhang, H., Go, Y.-M. & Jones, D. P. Mitochondrial thioredoxin-2/peroxiredoxin-3 system functions in parallel with mitochondrial GSH system in protection against oxidative stress. Arch. Biochem. Biophys. 465, 119–126 (2007).
Seelig, G. F., Simondsen, R. P. & Meister, A. Reversible dissociation of gamma-glutamylcysteine synthetase into two subunits. J. Biol. Chem. 259, 9345–9347 (1984).
Li, W., Li, Z., Yang, J. & Ye, Q. Production of glutathione using a bifunctional enzyme encoded by gshF from Streptococcus thermophilus expressed in Escherichia coli. J. Biotechnol. 154, 261–268 (2011).
Chen, W. W., Freinkman, E., Wang, T., Birsoy, K. & Sabatini, D. M. Absolute quantification of matrix metabolites reveals the dynamics of mitochondrial metabolism. Cell 166, 1324–1337.e11 (2016).
Slabbaert, J. R. et al. Shawn, the Drosophila homolog of SLC25A39/40, is a mitochondrial carrier that promotes neuronal survival. J. Neurosci. 36, 1914–1929 (2016).
Usaj, M. et al. TheCellMap.org: a web-accessible database for visualizing and mining the global yeast genetic interaction network. Genes Genomes Genet. 7, 1539–1549 (2017).
Luk, E., Carroll, M., Baker, M. & Culotta, V. C. Manganese activation of superoxide dismutase 2 in Saccharomyces cerevisiae requires MTM1, a member of the mitochondrial carrier family. Proc. Natl Acad. Sci. USA 100, 10353–10357 (2003).
Nilsson, R. et al. Discovery of genes essential for heme biosynthesis through large-scale gene expression analysis. Cell Metab. 10, 119–130 (2009).
Biederbick, A. et al. Role of human mitochondrial Nfs1 in cytosolic iron–sulfur protein biogenesis and iron regulation. Mol. Cell. Biol. 26, 5675–5687 (2006).
Mayr, J. A., Feichtinger, R. G., Tort, F., Ribes, A. & Sperl, W. Lipoic acid biosynthesis defects. J. Inherit. Metab. Dis. 37, 553–563 (2014).
Chen, Z. & Lash, L. H. Evidence for mitochondrial uptake of glutathione by dicarboxylate and 2-oxoglutarate carriers. J. Pharmacol. Exp. Ther. 285, 608–618 (1998).
Booty, L. M. et al. The mitochondrial dicarboxylate and 2-oxoglutarate carriers do not transport glutathione. FEBS Lett. 589, 621–628 (2015).
Kumar, C. et al. Glutathione revisited: a vital function in iron metabolism and ancillary role in thiol-redox control. EMBO J. 30, 2044–2056 (2011).
Rodríguez-Manzaneque, M. T., Tamarit, J., Bellí, G., Ros, J. & Herrero, E. Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol. Biol. Cell 13, 1109–1121 (2002).
Almusafri, F. et al. Clinical and molecular characterization of 6 children with glutamate–cysteine ligase deficiency causing hemolytic anemia. Blood Cells. Mol. Dis. 65, 73–77 (2017).
Wessel, D. & Flügge, U. I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143 (1984).
McAlister, G. C. et al. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem. 86, 7150–7158 (2014).
Tyanova, S. et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 (2016).
Birsoy, K. et al. An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell 162, 540–551 (2015).
Concordet, J.-P. & Haeussler, M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 46, W242–W245 (2018).
Shola, D. T. N., Yang, C., Han, C., Norinsky, R. & Peraza, R. D. in Mouse Genetics: Methods and Protocols (eds. Singh, S. R., Hoffman, R. M. & Singh, A.) 1–27 (Springer US, 2021).
Murgha, Y. E., Rouillard, J.-M. & Gulari, E. Methods for the preparation of large quantities of complex single-stranded oligonucleotide libraries. PLoS ONE 9, e94752 (2014).
Sadreyev, I. R., Ji, F., Cohen, E., Ruvkun, G. & Tabach, Y. PhyloGene server for identification and visualization of co-evolving proteins using normalized phylogenetic profiles. Nucleic Acids Res. 43, W154–W159 (2015).
Ruan, J. et al. TreeFam: 2008 update. Nucleic Acids Res. 36, D735–D740 (2008).
Thomas, P. D. et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13, 2129–2141 (2003).
Studer, G. et al. ProMod3—a versatile homology modelling toolbox. PLOS Comput. Biol. 17, e1008667 (2021).
Pebay-Peyroula, E. et al. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature 426, 39–44 (2003).
Robinson, A. J. & Kunji, E. R. S. Mitochondrial carriers in the cytoplasmic state have a common substrate binding site. Proc. Natl. Acad. Sci. 103, 2617–2622 (2006).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018).
Barbeira, A. N. et al. Integrating predicted transcriptome from multiple tissues improves association detection. PLoS Genet. 15, e1007889 (2019).
Zhou, D. et al. A unified framework for joint-tissue transcriptome-wide association and Mendelian randomization analysis. Nat. Genet. 52, 1239–1246 (2020).
Unlu, G. et al. Phenome-based approach identifies RIC1-linked Mendelian syndrome through zebrafish models, biobank associations and clinical studies. Nat. Med. 26, 98–109 (2020).
Aguet, F. et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017).
Acknowledgements
We thank all members of the Birsoy laboratory for helpful suggestions. Y.W. is supported by UL1 TR001866 from the National Center for Advancing Translational Sciences (NCATS, National Institutes of Health (NIH) Clinical and Translational Science Award (CTSA) programme. F.S.Y. and R.W. are supported by a Medical Scientist Training Program grant from the National Institute of General Medical Sciences of the NIH under award number T32GM007739 to the Weill Cornell/Rockefeller/Sloan Kettering Tri-Institutional MD-PhD Program. G.U. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-2431-21). E.R.G. was supported by NIH/NHGRI R01HG011138 and R35HG010718. H.L. is funded by NIH K99 DK128602-01. M.G.K. is a Scholar of the Leukemia and Lymphoma Society and is supported by the US NIH NIDDK R01-DK101989-01A1; NCI 1R01CA193842-01, 1R01CA193842-06A1, 5R01CA186702-07, 1R01DK1010989-06A1, R01HL135564 and R01CA225231-01; NYSTEM 0266-A121-4609, the Alex’s Lemonade Stand A Award. R.H. is supported by NIH/NCI Cancer Center Support Grant (P30 CA008748) and the Searle Scholars Program. We thank R. Vaughan, Director of Biostatistics at the Rockefeller University for his assistance with statistics used in this manuscript. K.B. is supported by the NIH/NCI (DP2 OD024174-01), NIH/NIDDK (R01 DK123323-01), Pershing Square Sohn Foundation and Mark Foundation Emerging Leader Award; and is a Searle and Pew-Stewart Scholar.
Author information
Authors and Affiliations
Contributions
K.B. conceived the project and K.B., Y.W., F.S.Y., X.G.Z., R.C.T. and R.W. designed the experiments. Y.W., F.S.Y., X.G.Z. and R.C.T. performed most of the experiments with the assistance of H.-W.Y., R.W. and Y.L. R.C.T. and R.W. designed and engineered GshF constructs. H.L. and M.G.K. characterized the mouse phenotypes. C.X., B.A. and R.H. assisted with the substrate mutant experiments. G.U. and E.R.G. performed the human genetic and transcriptome analysis. S.H. performed proteomics analysis. K.B. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
K.B. is scientific advisor to Nanocare Pharmaceuticals and Barer Institute.
Additional information
Peer review information Nature thanks Michael Murphy, Tracey Rouault and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 SLC25A39 protein levels are directly regulated by cellular GSH availability.
a, Gene enrichment analysis (Gorilla) for proteomics data from the immunopurified mitochondria of HeLa cells (top). Gene ontology analysis of proteins altered by BSO treatment (bottom). Log (FDR), log false discovery rate (bottom). Statistical significance was determined by Fisher’s exact or binomial distribution test. b, Immunoblot of SLC25A39, SLC25A12 and CS in HeLa cells treated with the indicated doses of BSO for 24 h. CS was used as the loading control. c, Immunoblot of indicated proteins in HEK-293T (top) and K562 (bottom) cells treated with BSO (1 mM;10 μM, respectively) for the indicated days. CS and GAPDH were used as loading controls. d, Immunofluorescence analysis of indicated proteins in HeLa cells treated with vehicle or BSO (1 mM) for 24 h. Micrographs are representative images. Scale bar, 10 μm (top). Immunofluorescence analysis of SLC25A39 in parental and SLC25A39 knockout HeLa cells treated with BSO (1 mM) and erastin (5 uM) for 24 h (bottom). Micrographs are representative images of three independent experiments. Scale bar, 20 μm. e, Relative abundance of SLC25A39, GCLM and SLC25A40 transcripts in HEK-293T cells treated with BSO (1 mM) for the indicated days using quantitative reverse transcription PCR (RT-qPCR), normalized to transcripts of β-ACTIN. Error bars represent mean ± s.d.; n = 3 biologically independent samples. Statistical significance was determined by one-way ANOVA followed by Bonferroni post-hoc analysis. f, Immunoblot of indicated proteins in HEK-293T SLC25A39 knockout cells expressing a vector control or SLC25A39 cDNA treated with BSO (1 mM) for the indicated times. GAPDH was used as the loading control. g, The design of 3xFLAG- SLC25A39-P2A-RFP construct (top). Immunoblot of indicated proteins in HEK-293T cells expressing 3xFLAG- SLC25A39-P2A-RFP treated with BSO (1 mM) and erastin (5 μM) for 48 h (bottom). \(\alpha \)-Tubulin was used as the loading control. h, Immunoblot of indicated proteins in HeLa cells treated with indicated doses of H2O2 for 24 h. CS and GAPDH were used as loading controls. i, Immunoblot of indicated proteins in HeLa cells treated with erastin (5 μM) or indicated doses of KI696 (NRF2 activator) for 24 h. GAPDH were used as loading controls. j, Immunoblot of indicated proteins in HeLa cells treated with erastin (5 μM) or indicated doses of RSL-3, an inhibitor of glutathione peroxidase 4 (GPX4), for 24 h. CS was used as a loading control. k, Immunoblot of SLC25A39 and ATF4 proteins in HeLa cells treated with indicated doses of mitoCDNB, diamide and BSO (1 mM)/erastin (5 μM) for 24 h. GAPDH was used as loading control. l, Immunoblot of indicated proteins in HeLa cells treated with erastin (5 μM) and co-treated with either GSH (10 mM), GSHee (10 mM), Trolox (50 μM) or Ferrostatin-1 (5 μM) for 48 h. CS was used as a loading control. GSHee, glutathione ethyl ester.
Extended Data Fig. 2 Mitochondrial GSH import is mediated by SLC25A39.
a, Volcano plots showing the fold change in mitochondrial metabolite abundance (log2) versus P values (-log) from K562 and HEK-293T SLC25A39 knockout cell lines expressing a vector control or SLC25A39 cDNA. Red data points highlight GSH and GSSG. The dotted line represents P = 0.05. Statistical significance was determined by multiple two-tailed unpaired t-tests with correction for multiple comparisons using the Holm-Šídák method. b, Immunoblot of indicated proteins in whole cell lysates and mitochondria isolated from HEK-293T (top) and HeLa (bottom) SLC25A39 knockout cells expressing a vector control or SLC25A39 cDNA. c, Mitochondrial GSH and GSSG abundance (top and middle panels) and total intracellular GSH levels in HEK-293T parental or SLC25A39 knockout cells expressing a vector control or SLC25A39 cDNA. Data are normalized by citrate synthase (CS) protein levels. d, Uptake of indicated concentrations of [13C2,15N]-GSH into mitochondria isolated from parental HEK-293T mitotag cells for 10 min. Data are normalized to the NAD+ abundance. e, Uptake of indicated concentrations of [13C2,15N]-GSH or GSH into mitochondria isolated from parental HEK-293T mitotag cells for 10 min. Data are normalized to the NAD+ abundance. f, Uptake of [13C2,15N]-GSH (5 mM, top) or [13C4,15N2]-GSSG (0.5 mM, bottom) into mitochondria isolated from HEK-293T SLC25A39 knockout cells expressing a vector control or SLC25A39 cDNA for 10 min. Data are normalized by citrate synthase (CS) protein levels. g, Uptake of [13C2,15N]-GSH (5 mM) into mitochondria isolated from HEK-293T SLC25A39 knockout cells expressing SLC25A39 cDNA for 10 min in the presence or absence of DTT (100 mM). Data are normalized by citrate synthase (CS) protein levels. h, Mitochondrial GSH abundance in HeLa SLC25A39 knockout cells expressing indicated cDNAs (top). Data are normalized by citrate synthase (CS) protein levels. Immunoblot of indicated proteins in HeLa SLC25A39 knockout cells expressing indicated cDNAs (bottom). α-Tubulin was used as a loading control. c, d, e, f, g, h, Bars represent mean ± s.d.; a, c, d, e, f, g, h, n = 3 biologically independent samples. Statistical significance in c, d, f, g and h was determined by one-way ANOVA followed by Bonferroni post-hoc analysis; a, e by two-tailed unpaired t-test.
Extended Data Fig. 3 SLC25A40, the mammalian paralog for SLC25A39 can compensate SLC25A39 loss.
a, Individual sgRNA scores of SLC25A40 and TXNRD2 from the CRISPR screens in indicated cell lines from Fig. 3b. b, CRISPR gene scores in indicated SLC25A39 knockout HEK-293T cells expressing a vector control or SLC25A39 cDNA. SLC25A40 data point is highlighted in blue (top). Top 15 scoring genes differentially required for the proliferation of HEK-293T SLC25A39 knockout cells (bottom). c, Phylogenetic tree of SLC25A39 homologs across model organisms. d, Top ranked (z-scores) co-evolved genes with SLC25A40 across species. e, Relative conservation of SLC25A40 and SLC25A39 in different species compared to their human homologs. f, Representative bright-field micrographs of Jurkat SLC25A39 knockout cells transduced with the indicated sgRNA at the end of the cell proliferation assay. Scale bar, 50 µm (right). g, Relative fold change in cell number of the indicated HEK-293T SLC25A39 knockout cells expressing a vector control or SLC25A39 cDNA transduced with the indicated sgRNAs. Cells were cultured for 4 days (mean ± SD, n = 3). Cell doublings were normalized to the average of the SLC25A39 knockout cells expressing SLC25A39 cDNA. Statistical significance was determined by one-way ANOVA followed by Bonferroni post-hoc analysis. h, Volcano plots of the fold change in mitochondrial metabolite abundance (log2) versus P values (-log) from HEK-293T SLC25A39 knockout cell line expressing a vector control or SLC25A40 cDNA. Red data points highlight GSH and GSSG. The dotted line represents P = 0.05. i, Immunoblot of indicated proteins in HEK-293T cells expressing an SLC25A40 cDNA in the presence or absence of BSO. CS and β-actin were used as loading controls.
Extended Data Fig. 4 Expression of S.thermophilus GshF can modulate cellular GSH levels in mammalian cells.
a, Mitochondrial abundance of GSH in HEK-293T SLC25A39 knockout cells expressing a vector control or SLC25A39 cDNA treated with GSH (5 mM) or GSHee (5 mM) for 24 h. Data are normalized by CS protein levels. b, Relative cell number of indicated SLC25A39 knockout Jurkat cells transduced with indicated sgRNAs. Cells were cultured for 4 days with or without GSHee (10 mM). Cell numbers were normalized to the average of the SLC25A39 knockout cells in each treatment condition. c, Immunofluorescence analysis of GshF (FLAG, red) and CS (green) in HeLa cells. Micrographs are representative images. Scale bar, 20 μm. d, Immunoblots of indicated proteins in HEK-293T cells expressing a vector control or GshF cDNA. GAPDH was used as the loading control. e, Schematic of engineered GshF construct for mammalian expression and the domains of the protein with GCL and GS function (top). Whole cell GSH abundance (bottom left) and fold change in cell number (log2) of HEK-293T cells expressing a vector control or engineered GshF cDNA treated with indicated BSO concentrations for 5 days (bottom right). f, Immunoblot analysis of indicated proteins in whole-cell lysates and mitochondria isolated from SLC25A39 knockout HEK-293T cells expressing a vector control, mito-GshF or SLC25A39 cDNA. g, Mitochondrial abundance of GSSG in HEK-293T SLC25A39 knockout cells transduced with the indicated cDNAs. Data are normalized by CS protein levels. a, b, e, g Bars represent mean ± s.d.; a, b, e, g n = 3 biologically independent samples. Statistical significance in a, g were determined by one-way ANOVA followed by Bonferroni post-hoc analysis; b, e were determined by two-way ANOVA followed by Bonferroni post-hoc analysis.
Extended Data Fig. 5 Mitochondrial GSH depletion sensitizes cells to BSO treatment.
a, Scheme of the in vitro sgRNA competition assay performed in HEK-293T SLC25A39 knockout cells expressing a vector control or SLC25A39 cDNA transduced with a pool of 5 control sgRNAs (sgControl, gray) and sgRNAs targeting TXNRD2 (pink) and SLC25A40 (green) (left). Differential guide scores in the indicated cell lines upon treatment with 20 μM BSO (right). Bars represent mean ± s.d. b, Synthetic lethal genetic interactions of mtm1 with other genes in S. cerevisiae.
Extended Data Fig. 6 Slc25a39 is essential for embryonic development and red cell differentiation in vivo.
a, Targeting scheme for Slc25a39 knockout mice. b, Gross appearance of E12.5 embryos of the indicated genotypes. Genotyping of Slc25a39 E12.5 embryos from heterozygous mating. PCR of wild type allele and targeted allele result in ~1,500bp and ~800bp bands, respectively (left). The number of viable pups with indicated genotypes is shown (right). c, Targeting scheme for Slc25a39 conditional knockout (Slc25a39fl/fl) mice in which two loxp sites were inserted in the indicated intronic regions. The resultant Slc25a39fl/fl mice were mated with erythroid-lineage specific Cre-recombinase (ErGFP-cre) mice to generate an erythroid-specific conditional knockout mice. d, The number of viable pups with indicated genotypes is shown resulting from the indicated mating. e, Representative images of H&E staining of fetal liver cells from E12.5 Slc25a39fl/fl and ErGFP-cre+/- Slc25a39fl/fl embryos. The bottom images are the boxed area of the top images. Arrows show that many haematopoietic cells in the fetal liver display nuclear fragmentation and cellular shrinkage. Bars, 50 μm (top panel), 12.5 μm (bottom panel). f, Percent of Prussian blue positive peripheral blood cells from indicated E12.5 embryos (top). Representative images of Prussian blue staining of indicated E12.5 whole embryo sections (bottom). Blue and orange arrows indicate blood cells in developing heart with negative or positive staining, respectively. Bar, 360 μm. g, Gating strategy and quantification of erythroblast populations at different differentiation stages using surface markers: Ter119 and CD44, and FSC (size). Percentage of cells of indicated populations among total fetal liver cells (left). Gating strategy (right). n = 3, ErGFP-cre+/- Slc25a39fl/fl; n = 4 Slc25a39fl/fl embryos. h, Profiling of cells of myeloid lineage (top), myeloid progenitor cells (middle) and lymphoid lineage (bottom). Data are presented as the population among total fetal liver cells. E12 embryos: n = 12 Slc25a39fl/fl or Slc25a39fl/-, n = 7 ErGFP-cre+/- Slc25a39fl/-, n = 4 ErGFP-cre+/- Slc25a39fl/fl. i, Relative RPKM values of SLC25A39, FTL and HBB genes in human erythroblasts at differing stages of terminal differentiation. f, g and h, Bars represent mean ± s.d.; statistical significance in f and h was determined by one-way ANOVA followed by Bonferroni post-hoc analysis; g was determined by two-tailed unpaired t-test.
Extended Data Fig. 7 Loss of SLC25A39/40 decreases the steady state levels of iron-sulfur containing proteins and phenocopies iron-sulfur cluster deficiency.
a, Comparison of proteomics data from BSO treated HeLa cells and SLC25A39/40 double knockout Jurkat cells. The criteria are indicated on the Venn diagram. b, Gene ontology analysis of significantly downregulated genes in SLC25A39/40 double knockout Jurkat cells. FDR, false discovery rate. c, Immunoblot of indicated proteins in the indicated HEK-293T SLC25A39 knockout cells expressing a vector control or SLC25A39 cDNA transduced with the indicated sgRNA. β-ACTIN was used as a loading control. d, Immunoblot of indicated proteins in indicated HEK-293T cells transduced with shRNA targeting either GFP as a control or NFS1 (two different shRNAs). Proteins were extracted from cells 6 days after transduction. GAPDH was used as a loading control. e, Schematic showing how the iron-sulfur cluster-containing protein LIAS enables OGDH and PDH activity by transferring lipoic acid as a cofactor to their enzyme complexes (left). Heatmap showing fold change in metabolite levels (log2) of the indicated cell lines relative to the average of those in SLC25A39 knockout cells complemented with SLC25A39 cDNA (middle). Metabolites were extracted 5 days after transduction with indicated sgRNAs. Values were normalized to the protein concentration of each cell line (left). Relative ratios of pyruvate to citrate metabolite levels of each indicated cell line from the metabolomics analysis in Fig. 4e (middle). Ratios were normalized to the average of the SLC25A39 knockout cells expressing SLC25A39 cDNA. Immunoblot with antibody against lipoic acid in the indicated Jurkat SLC25A39 knockout cells expressing a vector control or SLC25A39 cDNA transduced with the indicated sgRNA. Arrows indicate E2 complexes of PDH and OGDH containing lipoic acid (right). f, Representative bright-field micrographs of Jurkat SLC25A39 knockout cells transduced with the indicated sgRNA at the end of the cell proliferation assay. Scale bar, 50 µm. g, Relative cell number of the indicated Jurkat SLC25A39 knockout cells expressing a vector control or SLC25A39 cDNA transduced with the indicated sgRNA. Cells were cultured for 4 days with or without hemin (1 μM) and pyruvate (1 mM). Cell numbers were normalized to the average of the SLC25A39 knockout cells in each treatment condition. e, g, Bars represent mean ± s.d.; e, g, n = 3 biologically independent samples. Statistical significance in e was determined by one-way ANOVA followed by Bonferroni post-hoc analysis; g was determined by two-way ANOVA followed by Bonferroni post-hoc analysis.
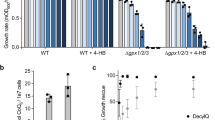
Extended Data Fig. 8 Expression of DIC/OGC does not complement the decrease of mitochondrial GSH availability in SLC25A39 knockout cells.
a, Uptake of [13C2,15N]-GSH into mitochondria isolated from HEK-293T SLC25A39 knockout cells expressing a vector control, SLC25A39 cDNA, SLC25A10 (DIC, mitochondrial dicarboxylate carrier) or SLC25A11 (OGC, mitochondrial oxoglutarate carrier) cDNA at 5 s and 10 min. Bars represent mean ± s.d.; n = 3 biologically independent samples. Statistical significance was determined by two-way ANOVA followed by Bonferroni post-hoc analysis. b, PrediXcan-based TWAS design in UK biobank and summary graph of SLC25A39-associated phenotypes. Bonferroni-adjusted p-values are reported.
Supplementary information
Supplementary Figure
This file contains Supplementary Fig. 1, the unprocessed Western blots and/or gels associated with the data presented in the figures and extended data figures.
Supplementary Table 1
Global analysis of mitochondrial proteome of HeLa cells in response to cellular GSH depletion. Statistical significance was determined by two-tailed unpaired t-test.
Supplementary Table 2
Metabolism focused CRISPR genetic screens in SLC25A39-knockout Jurkat and HEK 293T cells.
Supplementary Table 3
Whole-cell proteomic analysis of Jurkat cells in response to mitochondrial GSH depletion.
Source data
Rights and permissions
About this article
Cite this article
Wang, Y., Yen, F.S., Zhu, X.G. et al. SLC25A39 is necessary for mitochondrial glutathione import in mammalian cells. Nature 599, 136–140 (2021). https://doi.org/10.1038/s41586-021-04025-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-04025-w
This article is cited by
-
TREM2 macrophage promotes cardiac repair in myocardial infarction by reprogramming metabolism via SLC25A53
Cell Death & Differentiation (2024)
-
The pleiotropic functions of reactive oxygen species in cancer
Nature Cancer (2024)
-
Mitophagy mediated by BNIP3 and NIX protects against ferroptosis by downregulating mitochondrial reactive oxygen species
Cell Death & Differentiation (2024)
-
Mitochondrial proteome research: the road ahead
Nature Reviews Molecular Cell Biology (2024)
-
A CRISPRi/a screening platform to study cellular nutrient transport in diverse microenvironments
Nature Cell Biology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.