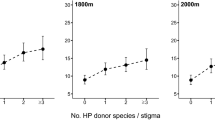
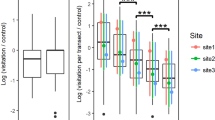
Abstract
Mechanisms that favour rare species are key to the maintenance of diverse communities1,2,3. One of the most critical tasks for conservation of flowering plant biodiversity is to understand how plant–pollinator interactions contribute to the maintenance of rare species4,5,6,7. Here we show that niche partitioning in pollinator use and asymmetric facilitation confer fitness advantage of rarer species in a biodiversity hotspot using phylogenetic structural equation modelling that integrates plant–pollinator and interspecific pollen transfer networks with floral functional traits. Co-flowering species filtered pollinators via floral traits, and rarer species showed greater pollinator specialization leading to higher pollination-mediated male and female fitness than more abundant species. When plants shared pollinator resources, asymmetric facilitation via pollen transport dynamics benefitted the rarer species at the cost of more abundant species, serving as an alternative diversity-promoting mechanism. Our results emphasize the importance of community-wide plant–pollinator interactions that affect reproduction for biodiversity maintenance.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
All data that support the findings of this study are included in this published article and its Supplementary Information files and source data files. Source data are provided with this paper.
Code availability
All software used in this study are provided in the Methods, Supplementary Information and the accompanying Reporting Summary.
References
Hubbell, S. P. The Unified Neutral Theory of Biodiversity and Biogeography (Princeton Univ. Press, 2001).
Wills, C. et al. Nonrandom processes maintain diversity in tropical forests. Science 311, 527–531 (2006).
Chesson, P. Mechanisms of maintenance of species diversity. Annu. Rev. Ecol. Evol. Syst. 31, 343–366 (2000).
Ollerton, J. Pollinator diversity: distribution, ecological function, and conservation. Annu. Rev. Ecol. Evol. Syst. 48, 353–376 (2017).
Vamosi, J. C. et al. Pollination decays in biodiversity hotspots. Proc. Natl Acad. Sci. USA 103, 956–961 (2006).
Bennett, J. M. et al. Land use and pollinator dependency drives global patterns of pollen limitation in the Anthropocene. Nat. Commun. 11, 3999 (2020).
Potts, S. G. et al. Global pollinator declines: trends, impacts and drivers. Trends Ecol. Evol. 25, 345–353 (2010).
Vamosi, J. C., Magallon, S., Mayrose, I., Otto, S. P. & Sauquet, H. Macroevolutionary patterns of flowering plant speciation and extinction. Annu. Rev. Plant Biol. 69, 685–706 (2018).
Ollerton, J., Winfree, R. & Tarrant, S. How many flowering plants are pollinated by animals? Oikos 120, 321–326 (2011).
Rodger, J. G. et al. 2021 Widespread vulnerability of plant seed production to pollinator decline. Sci. Adv. (in the press).
Pimm, S. L., Jones, H. L. & Diamond, J. On the risk of extinction. Am. Nat. 132, 757–785 (1988).
Sargent, R. D. & Ackerly, D. D. Plant–pollinator interactions and the assembly of plant communities. Trends Ecol. Evol. 23, 123–130 (2008).
Benadi, G. & Pauw, A. Frequency dependence of pollinator visitation rates suggests that pollination niches can allow plant species coexistence. J. Ecol. 106, 1892–1901 (2018).
Bruno, J. F., Stachowicz, J. J. & Bertness, M. D. Inclusion of facilitation into ecological theory. Trends Ecol. Evol. 18, 119–125 (2003).
Benadi, G., Bluthgen, N., Hovestadt, T. & Poethke, H. J. Population dynamics of plant and pollinator communities: stability reconsidered. Am. Nat. 179, 157–168 (2012).
Moeller, D. A. Facilitative interactions among plants via shared pollinators. Ecology 85, 3289–3301 (2004).
Bergamo, P. J., Susin Streher, N., Traveset, A., Wolowski, M. & Sazima, M. Pollination outcomes reveal negative density-dependence coupled with interspecific facilitation among plants. Ecol. Lett. 23, 129–139 (2020).
Barrett, S. C. H. The evolution of plant sexual diversity. Nat. Rev. Genet. 3, 274–284 (2002).
Ashman, T. L. & Arceo-Gómez, G. Toward a predictive understanding of the fitness costs of heterospecific pollen receipt and its importance in co-flowering communities. Am. J. Bot. 100, 1061–1070 (2013).
Moreira-Hernández, J. I. & Muchhala, N. Importance of pollinator-mediated interspecific pollen transfer for angiosperm evolution. Annu. Rev. Ecol. Evol. Syst. 50, 191–217 (2019).
Ashman, T. L. et al. Pollen limitation of plant reproduction: ecological and evolutionary causes and consequences. Ecology 85, 2408–2421 (2004).
Tur, C., Saez, A., Traveset, A. & Aizen, M. A. Evaluating the effects of pollinator-mediated interactions using pollen transfer networks: evidence of widespread facilitation in south Andean plant communities. Ecol. Lett. 19, 576–586 (2016).
Levin, D. A. & Anderson, W. W. Competition for pollinators between simultaneously flowering species. Am. Nat. 104, 455–467 (1970).
Ashman, T. L., Alonso, C., Parra-Tabla, V. & Arceo-Gómez, G. Pollen on stigmas as proxies of pollinator competition and facilitation: complexities, caveats and future directions. Ann. Bot. 125, 1003–1012 (2020).
Lloyd, D. G. Some reproductive factors affecting the selection of self-fertilization in plants. Am. Nat. 113, 67–79 (1979).
Sargent, R. D. & Otto, S. P. The role of local species abundance in the evolution of pollinator attraction in flowering plants. Am. Nat. 167, 67–80 (2006).
Adler, P. B., Fajardo, A., Kleinhesselink, A. R. & Kraft, N. J. B. Trait-based tests of coexistence mechanisms. Ecol. Lett. 16, 1294–1306 (2013).
Armbruster, W. S. The specialization continuum in pollination systems: diversity of concepts and implications for ecology, evolution and conservation. Funct. Ecol. 31, 88–100 (2017).
Minnaar, C., Anderson, B., de Jager, M. L. & Karron, J. D. Plant–pollinator interactions along the pathway to paternity. Ann. Bot. 123, 225–245 (2019).
Kantsa, A. et al. Disentangling the role of floral sensory stimuli in pollination networks. Nat. Commun. 9, 1041 (2018).
Fang, Q. & Huang, S. Q. A directed network analysis of heterospecific pollen transfer in a biodiverse community. Ecology 94, 1176–1185 (2013).
Baldwin, B. G. Origins of plant diversity in the California floristic province. Annu. Rev. Ecol. Evol. Syst. 45, 347–369 (2014).
Bascompte, J., Jordano, P., Melian, C. J. & Olesen, J. M. The nested assembly of plant–animal mutualistic networks. Proc. Natl Acad. Sci. USA 100, 9383–9387 (2003).
Thomson, J. D., Fung, H. F. & Ogilvie, J. E. Effects of spatial patterning of co-flowering plant species on pollination quantity and purity. Ann. Bot. 123, 303–310 (2019).
Rezende, E. L., Lavabre, J. E., Guimaraes, P. R., Jordano, P. & Bascompte, J. Non-random coextinctions in phylogenetically structured mutualistic networks. Nature 448, 925–928 (2007).
Song, C. L., Rohr, R. P. & Saavedra, S. Why are some plant–pollinator networks more nested than others? J. Anim. Ecol. 86, 1417–1424 (2017).
Hegland, S. J., Nielsen, A., Lazaro, A., Bjerknes, A. L. & Totland, O. How does climate warming affect plant–pollinator interactions? Ecol. Lett. 12, 184–195 (2009).
Ohlemuller, R. et al. The coincidence of climatic and species rarity: high risk to small-range species from climate change. Biol. Lett. 4, 568–572 (2008).
Arceo-Gómez, G., Kaczorowski, R. L. & Ashman, T.-L. A network approach to understanding patterns of coflowering in diverse communities. Int. J. Plant Sci. 179, 569–582 (2018).
Koski, M. H. et al. Plant–flower visitor networks in a serpentine metacommunity: assessing traits associated with keystone plant species. Arthropod Plant Interact. 9, 9–21 (2015).
Arceo-Gómez, G. et al. Patterns of among- and within-species variation in heterospecific pollen receipt: the importance of ecological generalization. Am. J. Bot. 103, 396–407 (2016).
Chao, A. et al. Rarefaction and extrapolation with Hill numbers: a framework for sampling and estimation in species diversity studies. Ecol. Monogr. 84, 45–67 (2014).
R Core Team. R: A Language and Environment for Statistical Computing, https://www.R-project.org/ (R Foundation for Statistical Computing, 2019).
Arceo-Gómez, G., Alonso, C., Ashman, T.-L. & Parra-Tabla, V. Variation in sampling effort affects the observed richness of plant–plant interactions via heterospecific pollen transfer: implications for interpretation of pollen transfer networks. Am. J. Bot. 105, 1601–1608 (2018).
Hayes, R. A., Cullen N., Kaczorowski R. L., O’Neill E. M. & Ashman T-L. A community-wide description and key of pollen from co-flowering plants of the serpentine seeps of Mclaughlin Reserve. Madrono (in the press).
Dafni, A. Pollination Ecology: a Practical Approach (Oxford Univ. Press, 1992).
McMurdie, P. J. & Holmes, S. Waste NOT, want not: why rarefying microbiome data is inadmissible. PLoS Comp. Biol. 10, e1003531 (2014).
Qian, H. & Jin, Y. An updated megaphylogeny of plants, a tool for generating plant phylogenies and an analysis of phylogenetic community structure. J. Plant Ecol. 9, 233–239 (2016).
Zanne, A. E. et al. Three keys to the radiation of angiosperms into freezing environments. Nature 506, 89–92 (2014).
Hinchliff, C. E. et al. Synthesis of phylogeny and taxonomy into a comprehensive tree of life. Proc. Natl Acad. Sci. USA 112, 12764–12769 (2015).
Paradis, E. & Schliep, K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528 (2019).
Michonneau, F., Brown, J. W. & Winter, D. J. rotl: an R package to interact with the Open Tree of Life data. Methods Ecol. Evol. 7, 1476–1481 (2016).
Revell, L. J. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223 (2012).
Le, S., Josse, J. & Husson, F. FactoMineR: an R package for multivariate analysis. J. Stat. Softw. 25, 1–18 (2008).
Dormann, C. F., Gruber, B. & Fruend, J. Introducing the bipartite package: analysing ecological networks. R News 8, 8–11 (2008).
Feinsinger, P., Spears, E. E. & Poole, R. W. A simple measure of niche breadth. Ecology 62, 27–32 (1981).
Horn, H. S. Measurement of "overlap" in comparative ecological studies. Am. Nat. 100, 419–424 (1966).
Almeida-Neto, M., Guimaraes, P., Guimaraes, P. R., Loyola, R. D. & Ulrich, W. A consistent metric for nestedness analysis in ecological systems: reconciling concept and measurement. Oikos 117, 1227–1239 (2008).
Csardi, G. & Nepusz, T. The igraph software package for complex network research. InterJournal 1695, 1–9 (2006).
Patefield, W. Algorithm AS 159: an efficient method of generating random R × C tables with given row and column totals. Appl. Stat. 30, 91–97 (1981).
Oksanen, J. et al. vegan: Community Ecology Package. R package version 2.5–5, https://CRAN.R-project.org/package=vegan (2019).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2016).
Bastian, M., Heymann, S. & Jacomy, M. Gephi: an open source software for exploring and manipulating networks. Presented at the Third international AAAI Conference on Weblogs and Social Media (2009).
Arceo-Gómez, G., Kaczorowski, R. L., Patel, C. & Ashman, T. L. Interactive effects between donor and recipient species mediate fitness costs of heterospecific pollen receipt in a co-flowering community. Oecologia 189, 1041–1047 (2019).
Keck, F., Rimet, F., Bouchez, A. & Franc, A. phylosignal: an R package to measure, test, and explore the phylogenetic signal. Ecol. Evol. 6, 2774–2780 (2016).
Orme, D. et al. caper: comparative analyses of phylogenetics and evolution in R. R package version 1.0.1, https://CRAN.R-project.org/package=caper (2018).
Barrett, S. C. H. The evolution of plant sexual diversity. Nat. Rev. Genet. 3, 274–284 (2002).
Fort, H., Vazquez, D. P. & Lan, B. L. Abundance and generalisation in mutualistic networks: solving the chicken-and-egg dilemma. Ecol. Lett. 19, 4–11 (2016).
Pinheiro, J., Bates, D., DebRoy, S., Sarkar, D. & R Core Team. nlme: linear and nonlinear mixed effects models. R package version 3.1-143, https://CRAN.R-project.org/package=nlme (2019).
Lefcheck, J. S. & Freckleton, R. piecewiseSEM: piecewise structural equation modelling in R for ecology, evolution, and systematics. Methods Ecol. Evol. 7, 573–579 (2015).
Fox, J. & Weisberg, S. An R companion to Applied Regression, 3rd edition (Sage, 2019).
Blüthgen, N., Menzel, F. & Blüthgen, N. Measuring specialization in species interaction networks. BMC Ecol. 6, 9 (2006).
Shipley, B. The AIC model selection method applied to path analytic models compared using a d-separation test. Ecology 94, 560–564 (2013).
van der Bijl, W. phylopath: easy phylogenetic path analysis in R. PeerJ 6, e4718 (2018).
Acknowledgements
We thank J. Baker, D. Chang, A. M. Arters, U. Meenakshinathan, K. Doleski, S. Barratt-Boyes, R. A. Ashman and M. Holden for assistance with stigma pollen identification, floral trait measurements and insect specimen processing; J. Rawlins, J. Pawelek, R. Androw and B. Coulter for insect identification; J. Hyland and V. Verdecia for logistic support at the Carnegie Museum of Natural History; McLaughlin field station staff for logistical support of field work; and the members of the Ashman, Wood and Turcotte laboratories for discussion. This work was supported by the National Science Foundation (DEB 1452386) to T.-L.A.
Author information
Authors and Affiliations
Contributions
T.-L.A. conceived the study. N.W. and T.-L.A. led the conceptual development. N.W. analysed the data. N.W., T.-L.A. and R.L.K. wrote the manuscript. N.W., T.-L.A., R.L.K. and G.A.-G. contributed to manuscript revisions. R.L.K., E.M.O., R.A.H., G.A.-G. and T.-L.A. collected the data.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Marcelo Aizen, Christopher Kaiser-Bunbury and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Community-wide plant–pollinator network.
a, Plant species (n = 79) coloured by families are arranged on the left according to phylogeny. The numbers of pollinator species that plants interacted with are shown as black bars and numbers within parentheses. b, Pollinator species (n = 416) are arranged along the top according to the size and similarity of plant assemblages that they interacted with. c, The observed numbers of interactions are denoted as frequency (‘Freq’) by the colour scale
Extended Data Fig. 2 Rarefaction shows that the majority of pollinator diversity was captured with our sampling intensity.
Rarefaction curves of each of the 79 plant species (a, b) that were observed for plant–pollinator interactions (Supplementary Table 1) and the 64 plant species (c, d) that were included in the phylogenetic structural equation models (Fig. 2). The observed number of pollinators is represented by the solid portion of each coloured line, whereas the dashed portion indicates extrapolation in the rarefaction analysis using the R package iNEXT42. Lines colours are randomly assigned. Pollinator diversity, especially Chao’s Shannon diversity (b, d), started to level off at the observed number of pollinators for most plant species, reflecting sufficient sampling to capture pollinator diversity
Extended Data Fig. 3 Floral trait variation and abundance.
a, Multivariate analysis (factor analysis of mixed data, FAMD) of 20 floral traits (Supplementary Table 3). Plant species (n = 73, abbreviated as the first two letters of genus and species names and coloured by plant family) are segregated along the first two dimensions, representing mainly size-related and other (shape/colour/inflorescence) floral traits, respectively. These traits vary independently from species floral abundance (symbol size). b, Species rarity based on floral abundance (log-transformed) was correlated with rarity based on occurrence in the number of surveyed plots (see Methods, two-sided Pearson’s correlation test, r = 0.64, t = 6.9, d.f. = 70, P = 1.8 × 10−9). The 95% confidence intervals of the mean are shown
Extended Data Fig. 4 Multivariate analysis of floral traits associated with pollinator attraction.
a, b, In the first four dimensions of the factor analysis of mixed data (FAMD), the centroid of each category within a qualitative trait is indicated, with symbol shape representing different qualitative traits. Quantitative traits are represented by arrows. Individual plant species (n = 73) are shown in the background with colours indicating plant families and symbol sizes indicating floral abundances (Extended Data Fig. 3). c, The traits that contributed to ≥15% of variation of the first three dimensions are highlighted in colour
Extended Data Fig. 5 Multivariate analysis of floral traits associated with male organ.
a, b, In the first four dimensions of the factor analysis of mixed data (FAMD), the centroid of each category within a qualitative trait is indicated, with symbol shape representing different qualitative traits. Quantitative traits are represented by arrows. Individual plant species (n = 73) are shown in the background with colours indicating plant families and symbol sizes indicating floral abundances (Extended Data Fig. 3). c, The traits that contributed to ≥15% of variation of the first three dimensions are highlighted in colour
Extended Data Fig. 6 Multivariate analysis of floral traits associated with female organ.
a, b, In the first four dimensions of the factor analysis of mixed data (FAMD), the centroid of each category within a qualitative trait is indicated, with symbol shape representing different qualitative traits. Quantitative traits are represented by arrows. Individual plant species (n = 73) are shown in the background with colours indicating plant families and symbol sizes indicating floral abundances (Extended Data Fig. 3). c, The traits that contributed to ≥15% of variation of the first four dimensions are highlighted in colour
Extended Data Fig. 7 Pollen transfer network.
The network was constructed based on pollen deposited on 54 stigmas of 66 individual plant species (Supplementary Table 5). Plant species (nodes) are abbreviated as the first two letters of genus and species names (Supplementary Table 3), with unidentified species denoted with ‘U’. Node size indicates the number of flowering plant species that pollen is received from, and node colour darkness indicates the number of flowering plant species that pollen is donated to. That is, larger nodes represent better recipients and darker nodes better donors. Arrows and their sizes indicate the direction and amount (counts) of pollen transfer, respectively
Extended Data Fig. 8 Validation of fractional identity approach and rarefaction of pollen received by stigmas.
a, There was a strong relationship between heterospecific pollen (HP) richness when fractionally identified pollen grains were excluded (y-axis, ‘no ambiguity’) and included (x-axis, ‘fractional’): n = 66 plant species, general linear model, slope = 0.73, t = 17.1, P < 2 × 10−9. The dotted 95% confidence intervals of the mean are shown. b, c, Rarefaction analysis using the R package iNEXT42 showed that the majority of pollen species richness (b) and Chao’s Shannon diversity (c) were captured by the sampled styles (n = 54 on average) for each plant species (n = 66, coloured lines). The observed (solid) and extrapolated (dashed) portion of each rarefaction line are indicated
Extended Data Fig. 9 Phylogenetic structural equation models (PSEMs).
a–h, The PSEMs considered pollinator niche partitioning, asymmetric facilitation, pollination assurance and numeric assurance (orange arrows). Pollination assurance and numeric assurance are collectively referred to as automatic assurances. i, Model fitting and nested model selection used the R packages piecewiseSEM70 and phylopath74. Sample size was 64 plant species. df and P, degree of freedom and P value of the two-sided Fisher’s C statistic; AIC, the Akaike’s information criterion; CICc, the C statistic information criterion corrected for small sample sizes; w, CICc weights. Standardized regression coefficients of paths and model averaging are in Supplementary Table 6.
Supplementary information
Supplementary Information
This file contains Supplementary Methods, Supplementary Notes and Supplementary References.
Supplementary Table 1
Plant–pollinator interactions observed across sites and years.
Supplementary Table 2
Plant–pollinator network metrics and comparisons to null models.
Supplementary Table 3
Species-level floral traits.
Supplementary Table 4
Phylogenetic signals of plant–pollinator interactions, interspecific pollen transfer network metrics and floral traits.
Supplementary Table 5
Stigma pollen data.
Supplementary Table 6
Standardized regression coefficients and model averaging of phylogenetic structural equation models.
Rights and permissions
About this article
Cite this article
Wei, N., Kaczorowski, R.L., Arceo-Gómez, G. et al. Pollinators contribute to the maintenance of flowering plant diversity. Nature 597, 688–692 (2021). https://doi.org/10.1038/s41586-021-03890-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03890-9
This article is cited by
-
Genetic basis of nectar guide trichome variation between bumblebee- and self-pollinated monkeyflowers (Mimulus): role of the MIXTA-like gene GUIDELESS
BMC Plant Biology (2024)
-
Mutualisms weaken the latitudinal diversity gradient among oceanic islands
Nature (2024)
-
ALLENE OXIDE SYNTHASE (AOS) induces petal senescence through a novel JA-associated regulatory pathway in Arabidopsis
Physiology and Molecular Biology of Plants (2024)
-
Landfill fire impact on bee health: beneficial effect of dietary supplementation with medicinal plants and probiotics in reducing oxidative stress and metal accumulation
Environmental Science and Pollution Research (2023)
-
The interplay between scale, pollination niche and floral attractiveness on density-dependent plant–pollinator interactions
Oecologia (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.