Abstract
Signals from sympathetic neurons and immune cells regulate adipocytes and thereby contribute to fat tissue biology. Interactions between the nervous and immune systems have recently emerged as important regulators of host defence and inflammation1,2,3,4. Nevertheless, it is unclear whether neuronal and immune cells co-operate in brain–body axes to orchestrate metabolism and obesity. Here we describe a neuro-mesenchymal unit that controls group 2 innate lymphoid cells (ILC2s), adipose tissue physiology, metabolism and obesity via a brain–adipose circuit. We found that sympathetic nerve terminals act on neighbouring adipose mesenchymal cells via the β2-adrenergic receptor to control the expression of glial-derived neurotrophic factor (GDNF) and the activity of ILC2s in gonadal fat. Accordingly, ILC2-autonomous manipulation of the GDNF receptor machinery led to alterations in ILC2 function, energy expenditure, insulin resistance and propensity to obesity. Retrograde tracing and chemical, surgical and chemogenetic manipulations identified a sympathetic aorticorenal circuit that modulates ILC2s in gonadal fat and connects to higher-order brain areas, including the paraventricular nucleus of the hypothalamus. Our results identify a neuro-mesenchymal unit that translates cues from long-range neuronal circuitry into adipose-resident ILC2 function, thereby shaping host metabolism and obesity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Source data for quantifications shown in all graphs plotted in figures and extended data figures are available in the online version of the paper. The datasets generated in this study are also available from the corresponding author upon reasonable request. The RNA-seq datasets analysed are publicly available in the Gene Expression Omnibus repository with the accession numbers GSE179546 and GSE179551 for MSCs and ILC2s, respectively. Source data are provided with this paper.
References
Godinho-Silva, C., Cardoso, F. & Veiga-Fernandes, H. Neuro-immune cell units: a new paradigm in physiology. Annu. Rev. Immunol. 37, 19–46 (2019).
Huh, J. R. & Veiga-Fernandes, H. Neuroimmune circuits in inter-organ communication. Nat. Rev. Immunol. 20, 217–228 (2020).
Veiga-Fernandes, H. & Artis, D. Neuronal-immune system cross-talk in homeostasis. Science 359, 1465–1466 (2018).
Chu, C., Artis, D. & Chiu, I. M. Neuro-immune interactions in the tissues. Immunity 52, 464–474 (2020).
Bartness, T. J., Vaughan, C. H. & Song, C. K. Sympathetic and sensory innervation of brown adipose tissue. Int. J. Obes. 34 (Suppl. 1), S36–S42 (2010).
Morrison, S. F. Central neural control of thermoregulation and brown adipose tissue. Auton. Neurosci. 196, 14–24 (2016).
Zeng, W. et al. Sympathetic neuro-adipose connections mediate leptin-driven lipolysis. Cell 163, 84–94 (2015).
Moro, K. et al. Innate production of TH2 cytokines by adipose tissue-associated c-Kit+Sca-1+ lymphoid cells. Nature 463, 540–544 (2010).
Brestoff, J. R. et al. Group 2 innate lymphoid cells promote beiging of white adipose tissue and limit obesity. Nature 519, 242–246 (2015).
Hams, E. et al. The helminth T2 RNase ω1 promotes metabolic homeostasis in an IL-33- and group 2 innate lymphoid cell-dependent mechanism. FASEB J. 30, 824–835 (2016).
Lee, M. W. et al. Activated type 2 innate lymphoid cells regulate beige fat biogenesis. Cell 160, 74–87 (2015).
Molofsky, A. B. et al. Innate lymphoid type 2 cells sustain visceral adipose tissue eosinophils and alternatively activated macrophages. J. Exp. Med. 210, 535–549 (2013).
Vivier, E. et al. Innate lymphoid cells: 10 years on. Cell 174, 1054–1066 (2018).
Arighi, E., Borrello, M. G. & Sariola, H. RET tyrosine kinase signaling in development and cancer. Cytokine Growth Factor Rev. 16, 441–467 (2005).
Mulligan, L. M. RET revisited: expanding the oncogenic portfolio. Nat. Rev. Cancer 14, 173–186 (2014).
Fonseca-Pereira, D. et al. The neurotrophic factor receptor RET drives haematopoietic stem cell survival and function. Nature 514, 98–101 (2014).
Ibiza, S. et al. Glial-cell-derived neuroregulators control type 3 innate lymphoid cells and gut defence. Nature 535, 440–443 (2016).
Patel, A. et al. Differential RET signaling pathways drive development of the enteric lymphoid and nervous systems. Sci. Signal. 5, ra55 (2012).
Veiga-Fernandes, H. et al. Tyrosine kinase receptor RET is a key regulator of Peyer’s patch organogenesis. Nature 446, 547–551 (2007).
Kahn, B. B. & Flier, J. S. Obesity and insulin resistance. J. Clin. Invest. 106, 473–481 (2000).
Qiu, Y. et al. Eosinophils and type 2 cytokine signaling in macrophages orchestrate development of functional beige fat. Cell 157, 1292–1308 (2014).
Morrison, S. F., Madden, C. J. & Tupone, D. Central neural regulation of brown adipose tissue thermogenesis and energy expenditure. Cell Metab. 19, 741–756 (2014).
Sutton, A. K., Myers, M. G., Jr & Olson, D. P. The role of PVH circuits in leptin action and energy balance. Annu. Rev. Physiol. 78, 207–221 (2016).
Mahlakõiv, T. et al. Stromal cells maintain immune cell homeostasis in adipose tissue via production of interleukin-33. Sci. Immunol. 4, eaax0416 (2019).
Rana, B. M. J. et al. A stromal cell niche sustains ILC2-mediated type-2 conditioning in adipose tissue. J. Exp. Med. 216, 1999–2009 (2019).
Spallanzani, R. G. et al. Distinct immunocyte-promoting and adipocyte-generating stromal components coordinate adipose tissue immune and metabolic tenors. Sci. Immunol. 4, eaaw3658 (2019).
Ding, X. et al. IL-33-driven ILC2/eosinophil axis in fat is induced by sympathetic tone and suppressed by obesity. J. Endocrinol. 231, 35–48 (2016).
Moriyama, S. et al. β2-adrenergic receptor-mediated negative regulation of group 2 innate lymphoid cell responses. Science 359, 1056–1061 (2018).
Andersen, C. J., Murphy, K. E. & Fernandez, M. L. Impact of obesity and metabolic syndrome on immunity. Adv. Nutr. 7, 66–75 (2016).
Guilherme, A., Henriques, F., Bedard, A. H. & Czech, M. P. Molecular pathways linking adipose innervation to insulin action in obesity and diabetes mellitus. Nat. Rev. Endocrinol. 15, 207–225 (2019).
Zhuo, L. et al. hGFAP-cre transgenic mice for manipulation of glial and neuronal function in vivo. Genesis 31, 85–94 (2001).
Roesch, K. et al. The transcriptome of retinal Müller glial cells. J. Comp. Neurol. 509, 225–238 (2008).
de Boer, J. et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol. 33, 314–325 (2003).
Schlenner, S. M. et al. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity 32, 426–436 (2010).
Nussbaum, J. C. et al. Type 2 innate lymphoid cells control eosinophil homeostasis. Nature 502, 245–248 (2013).
Savitt, J. M., Jang, S. S., Mu, W., Dawson, V. L. & Dawson, T. M. Bcl-x is required for proper development of the mouse substantia nigra. J. Neurosci. 25, 6721–6728 (2005).
Hinoi, E. et al. The sympathetic tone mediates leptin’s inhibition of insulin secretion by modulating osteocalcin bioactivity. J. Cell Biol. 183, 1235–1242 (2008).
Mombaerts, P. et al. RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68, 869–877 (1992).
Cao, X. et al. Defective lymphoid development in mice lacking expression of the common cytokine receptor gamma chain. Immunity 2, 223–238 (1995).
Smith-Hicks, C. L., Sizer, K. C., Powers, J. F., Tischler, A. S. & Costantini, F. C-cell hyperplasia, pheochromocytoma and sympathoadrenal malformation in a mouse model of multiple endocrine neoplasia type 2B. EMBO J. 19, 612–622 (2000).
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010).
Sciolino, N. R. et al. Recombinase-dependent mouse lines for chemogenetic activation of genetically defined cell types. Cell Rep. 15, 2563–2573 (2016).
Buch, T. et al. A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat. Methods 2, 419–426 (2005).
Almeida, A. R. et al. RET/GFRα signals are dispensable for thymic T cell development in vivo. PLoS One 7, e52949 (2012).
Enomoto, H. et al. GFR alpha1-deficient mice have deficits in the enteric nervous system and kidneys. Neuron 21, 317–324 (1998).
Rossi, J. et al. Retarded growth and deficits in the enteric and parasympathetic nervous system in mice lacking GFR alpha2, a functional neurturin receptor. Neuron 22, 243–252 (1999).
Nishino, J. et al. GFR alpha3, a component of the artemin receptor, is required for migration and survival of the superior cervical ganglion. Neuron 23, 725–736 (1999).
Godinho-Silva, C. et al. Light-entrained and brain-tuned circadian circuits regulate ILC3s and gut homeostasis. Nature 574, 254–258 (2019).
Botta, P. et al. An amygdala circuit mediates experience-dependent momentary arrests during exploration. Cell 183, 605–619.e22 (2020).
Pereira, M. M. et al. A brain-sparing diphtheria toxin for chemical genetic ablation of peripheral cell lineages. Nat. Commun. 8, 14967 (2017).
Zhu, H. & Roth, B. L. Silencing synapses with DREADDs. Neuron 82, 723–725 (2014).
Leggett, R. M., Ramirez-Gonzalez, R. H., Clavijo, B. J., Waite, D. & Davey, R. P. Sequencing quality assessment tools to enable data-driven informatics for high throughput genomics. Front. Genet. 4, 288 (2013).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
McCarthy, D. J., Chen, Y. & Smyth, G. K. Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297 (2012).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Ebihara, T. & Taniuchi, I. Transcription factors in the development and function of group 2 innate lymphoid cells. Int. J. Mol. Sci. 20, 1377 (2019).
Grieger, J. C., Choi, V. W. & Samulski, R. J. Production and characterization of adeno-associated viral vectors. Nat. Protocols 1, 1412–1428 (2006).
Huang, X. et al. AAV2 production with optimized N/P ratio and PEI-mediated transfection results in low toxicity and high titer for in vitro and in vivo applications. J. Virol. Methods 193, 270–277 (2013).
Paxinos, G. & Franklin, K. B. J. The Mouse Brain in Stereotaxic Coordinates 2nd edition (Academic, 2001).
Huang, J. et al. A cationic near infrared fluorescent agent and ethyl-cinnamate tissue clearing protocol for vascular staining and imaging. Sci. Rep. 9, 521 (2019).
Acknowledgements
We thank the Vivarium, Flow Cytometry, Histopathology, Molecular Biology and Hardware platforms at the Champalimaud Centre for the Unknown. We thank Congento LISBOA-01-0145-FEDER-022170, co-financed by FCT (Portugal) and Lisboa2020, under the PORTUGAL2020 agreement (European Regional Development Fund). pAAV-Ef1a-mCherry-IRES-Cre was a gift from K. Deisseroth. PRV-614 (PRV-Bartha) was a gift from L. Enquist and E. Engel. F.C., C.G.-S., and R.G.D. were supported by Fundação para a Ciência e Tecnologia (FCT), Portugal. R.G.J.K.W. is supported by a Marie Skłodowska-Curie Individual fellowship (European Commission, 799810-TOPNIN), a Cancer Research Institute/Irvington Postdoctoral Fellowship and a Postdoctoral Junior Leader fellowship from la Caixa Foundation, ID100010434; LCF/BQ/PR20/11770004. H.V.-F. is supported by ERC (647274), EU, The Paul G. Allen Frontiers Group, US, and FCT, Portugal.
Author information
Authors and Affiliations
Contributions
F.C. designed, performed and analysed the experiments shown in Fig. 1–4 and Extended Data Fig. 1–9. R.G.J.K.W. performed the clearing and imaging in Fig. 1a, and the viral tracing and manipulation experiments in Fig. 4 and Extended Data Fig. 8. C.G.-S. and R.G.D. performed the electroablation surgeries in Extended Data Fig. 8. H.R. provided assistance for the experiments shown in Figs. 1–4. A.I.D. and I. M. provided technical help for the experiment shown in Fig. 1c. J.A.d.S. provided help for the experiment in Extended Data Fig. 2d, e. H.V.-F. supervised the work, planned the experiments and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks John Horn and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Gating strategy for ILC2s and MSCs.
a, ILC2s were defined as: live CD45+Lin−Thy1.2+Sca-1+KLRG1+ (lineage comprised CD3ε, CD8α, TCRβ, TCRγδ, CD19, GR1, CD11c, CD11b and TER119). b, Stroma cells were defined as: live PDGFRA+ MSCs (CD45–CD31–PDGFRA+gp38+SCa-1+), PDGFRA− MSCs (CD45−CD31−PDGFRA−gp38+), and endothelial cells (CD45−CD31+).
Extended Data Fig. 2 Sympathetic nervous system in the GAT and ILC2 function.
a, GAT. Red, sympathetic nerve fibres (TH); Green, endothelial cells (CD31). Scale bar, 300 μm. b, GAT ILC2-derived Met-enk after 6-OHDA administration. n = 5. c, CD4 T cells and TH+ CD4 T cells after 6-OHDA administration. n = 4. d, e, Distance covered by mice in the open field test. n = 3. Scale bar, 10 cm. f, GAT ILC2-derived Met-enk after clenbuterol administration. n = 5. g, Heatmap of Adrb1, Adrb2, and Adrb3 expression (read counts (fragments per kilobase of transcript per million mapped reads; FPKM) on ILC2s, n = 4). h, GAT ILC2s after CNO administration. R26/3Dfl n = 4, R26Th n = 4, and R26/3DTh n = 5. i, Heatmap of Adrb1, Adrb2, and Adrb3 expression (read counts (transcripts per million; TPM) on MSCs, n = 4). j, Gdnf expression after in vitro stimulation of MSCs, n = 3. Data are representative of three independent experiments. n represents biologically independent animals. Data are presented as mean ± s.e.m. Two-tailed unpaired Welch’s t-test. *P < 0.05; ***P < 0.005; ns, not significant
Extended Data Fig. 3 Sympathetic regulation of GAT MSCs.
a, Heatmap of genes upregulated in MSCs upon 6-OHDA administration. Vehicle n = 4, 6-OHDA n = 5. b, GAT Il33 expression after 6-OHDA treatment. n = 5. c, GAT Il33 expression after clenbuterol administration. n = 5. d, MSC-derived Il33 after 6-OHDA and clenbuterol administration. n = 6. e, MSC-derived Il25 after 6-OHDA and clenbuterol administration. n = 6. f, GDNF on MSCs. Adrb2fl n = 6, Adrb2ΔPdgfra n = 4. Data are representative of three independent experiments. n represents biologically independent animals. Data are presented as mean ± s.e.m. Two-tailed unpaired Welch’s t-test. ns, not significant
Extended Data Fig. 4 RET signals do not affect ILC2 differentiation and activation genes.
a, b, Heatmaps showing log(raw counts) of ILC2-related genes in ILC2s from Retfl (n = 4) and RetΔVav1 (n = 5) mice (a) and Rag1−/−RetWT (n = 3) and Rag1−/−RetMEN2B (n = 3) mice (b)
Extended Data Fig. 5 ILC2-autonomous RET signals control type 2 innate cytokines in the GAT.
a–c, GAT ILC2 function. a, Gfra1−/− fetal liver chimeras. n = 5. b, Gfra2+/+, n = 10; Gfra2−/−, n = 5. c, Gfra+/+, n = 8; Gfra3−/−, n = 8. d, RetΔVav1 mixed BM chimeras scheme. e, GAT ILC2 from RetΔVav1 mixed BM chimeras. Retfl n = 6; RetΔVav1 n = 7. f, ILC2s from Rag1−/−RetWT (n = 6) and Rag1−/−RetΔIl5 (n = 6) mice. g, ILC2s from RetWT (n = 10) and RetΔIl5 (n = 8) mice. h, RetΔIl5 mixed bone marrow (BM) chimeras scheme. i, GAT ILC2 from RetΔIl5 mixed BM chimeras. RetWT n = 4, RetΔIl5 n = 4. j, GAT ILC2s in Rag1−/−RetMEN2B BM chimeras. Rag1−/−RetWT n = 5, Rag1−/−RetMEN2B n = 6. k, RetMEN2B mixed BM chimeras scheme. l, Mixed BM chimeras. Rag1−/−RetWT n = 6, Rag1−/−RetMEN2B n = 7. Data are representative of three independent experiments. n represents biologically independent animals. Data are presented as mean ± s.e.m. Two-tailed unpaired Welch’s t-test. *P < 0.05; **P < 0.01; ***P < 0.005; ns, not significant
Extended Data Fig. 6 ILC2-intrinsic RET signalling is sufficient to control adipocyte physiology and obesity.
a, GAT ILC2s after 6-OHDA administration. RetWT n = 8 and RetΔIl5 n = 7. b, Weight gain during 16 weeks of HFD regimen. Rag1−/−RetWT n = 4, Rag1−/−RetΔIl5 n = 5. c, Intestinal lamina propria ILC3s. Rag1−/−RetWT n = 5, Rag1−/−RetΔIl5 n = 5. d, Weight gain during 16 weeks of HFD regimen. RetWT n = 5, RetΔIl5 n = 5. e, Frequency of ILC2 and ILC3 in Thy+Lin− lymphocytes from ILC2-chimeric mice after HFD. Each bar represents one mouse. n = 4. f, Total GAT RNA expression of Ucp1, Cox8b and Cidea. n = 5. Data are presented as mean ± s.e.m. Two-tailed unpaired Welch’s t-test (a, c, e); repeated measures ANOVA corrected for multiple comparisons with the Benjamini, Krieger and Yekutieli procedure (b, d); and Mann–Whitney test (f). *P < 0.05; **P < 0.01; ns, not significant
Extended Data Fig. 7 RET signals control adipose tissue energy expenditure.
Total RNA expression of adipose tissue-related genes in GAT. a, Retfl n = 4; RetΔVav1 n = 6. b, Rag1−/−RetWT n = 4; Rag1−/−RetMEN2B n = 5. c, GAT co-cultures scheme. d, GAT co-cultures with ILC2 and GDNF. Data are presented as mean ± s.e.m. Two-sided Mann–Whitney test. *P < 0.05; ns, not significant
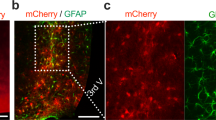
Extended Data Fig. 8 An aorticorenal–adipose circuit connects to the brain.
a, DRG at thoracic 13 (T13) level. Green, viral tracing (VT); red, TH. Scale bar, 100 μm. b, Left, brain atlas schemes of coronal sections. Right, polysynaptic tracing from the GAT corresponding to the highlighted areas on the left. c, Left, brain atlas schemes of coronal section. Right, polysynaptic tracing from the ARG corresponding to the highlighted areas on the left. b, c, Central amygdala (CA), zona incerta (ZI), periaqueductal grey (PAG), subcoeruleus nucleus (SubCD). Scale bars, 200 μm. d, Electrolytic lesion of the PVH. Scale bar, 500 μm. e, GAT ILC2s in PVH-ablated mice. Sham n = 5; PVH-ablated n = 6. f, GAT Il33 expression in AAV(4D) mice compared to contralateral controls after CNO administration. n = 5. g, GAT Il33 expression in AAV(3D) mice compared to contralateral control after CNO administration. n = 4. h, Scheme of combinatorial viral approach. The ARG was injected with an AAV carrying a Cre construct (AAV-Cre). Next, the GAT was injected with a Cre-inducible AAV(3Dfl). AAV(Cre) n = 7 and AAV(Cre)+AAV(3Dfl) n = 6. Data are representative of three independent experiments. n represents biologically independent animals. Data are presented as mean ± s.e.m. Two-tailed unpaired t-test (e); two-tailed Mann–Whitney test (f, g); two-tailed unpaired Welch’s t-test (h). *P < 0.05; **P < 0.01; ns, not significant
Extended Data Fig. 9 A sympathetic aorticorenal–adipose circuit connects to the brain and regulates ILC2s.
GAT neuro-mesenchymal units translate sympathetic cues into neurotrophic factor expression. In turn, neurotrophic factors control adipose ILC2 function via the neuroregulatory receptor RET, shaping the host metabolism, energy expenditure and obesity. SNS, sympathetic nervous system.
Supplementary information
Source data
Rights and permissions
About this article
Cite this article
Cardoso, F., Klein Wolterink, R.G.J., Godinho-Silva, C. et al. Neuro-mesenchymal units control ILC2 and obesity via a brain–adipose circuit. Nature 597, 410–414 (2021). https://doi.org/10.1038/s41586-021-03830-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03830-7
This article is cited by
-
Control of lipolysis by a population of oxytocinergic sympathetic neurons
Nature (2024)
-
Pathophysiological functions of semaphorins in the sympathetic nervous system
Inflammation and Regeneration (2023)
-
Central regulation of stress-evoked peripheral immune responses
Nature Reviews Neuroscience (2023)
-
Locally sourced: site-specific immune barriers to metastasis
Nature Reviews Immunology (2023)
-
Senescent immune cells accumulation promotes brown adipose tissue dysfunction during aging
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.