Abstract
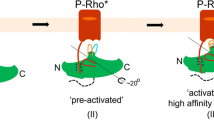
G-protein-coupled receptor (GPCR) kinases (GRKs) selectively phosphorylate activated GPCRs, thereby priming them for desensitization1. Although it is unclear how GRKs recognize these receptors2,3,4, a conserved region at the GRK N terminus is essential for this process5,6,7,8. Here we report a series of cryo-electron microscopy single-particle reconstructions of light-activated rhodopsin (Rho*) bound to rhodopsin kinase (GRK1), wherein the N terminus of GRK1 forms a helix that docks into the open cytoplasmic cleft of Rho*. The helix also packs against the GRK1 kinase domain and stabilizes it in an active configuration. The complex is further stabilized by electrostatic interactions between basic residues that are conserved in most GPCRs and acidic residues that are conserved in GRKs. We did not observe any density for the regulator of G-protein signalling homology domain of GRK1 or the C terminus of rhodopsin. Crosslinking with mass spectrometry analysis confirmed these results and revealed dynamic behaviour in receptor-bound GRK1 that would allow the phosphorylation of multiple sites in the receptor tail. We have identified GRK1 residues whose mutation augments kinase activity and crosslinking with Rho*, as well as residues that are involved in activation by acidic phospholipids. From these data, we present a general model for how a small family of protein kinases can recognize and be activated by hundreds of different GPCRs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data needed to evaluate the conclusions in the paper are presented in the paper and/or the Supplementary Materials. Additional data related to this paper are available upon reasonable request from the authors. The structures of the four Rho*–GRK1 complexes (Rho*–GRK1, Rho*–GRK1(S5E/EE), Rho*–GRK1(S5E/EE)–Fab1, and Rho*–GRK1(S5E/EE)–Fab6) and their associated data have been deposited into the Protein Data Bank under accession codes 7MT9, 7MT8, 7MTA, and 7MTB, and the Electron Microscopy Data Bank under accession codes EMD-23978, EMD-23977, EMD-23979, and EMD-23980, respectively. CLMS data have been deposited to the ProteomeXchange Database (http://www.proteomexchange.org/) via the PRIDE70 partner repository with dataset identifier PXD019215.
References
Gurevich, E. V., Tesmer, J. J., Mushegian, A. & Gurevich, V. V. G protein-coupled receptor kinases: more than just kinases and not only for GPCRs. Pharmacol. Ther. 133, 40–69 (2012).
Cato, M. C. et al. The open question of how GPCRs interact with GPCR kinases (GRKs). Biomolecules 11, 447 (2021).
Komolov, K. E. et al. Structural and functional analysis of a β2-adrenergic receptor complex with GRK5. Cell 169, 407–421.e16 (2017).
He, Y. et al. Molecular assembly of rhodopsin with G protein-coupled receptor kinases. Cell Res. 27, 728–747 (2017).
Beautrait, A. et al. Mapping the putative G protein-coupled receptor (GPCR) docking site on GPCR kinase 2: insights from intact cell phosphorylation and recruitment assays. J. Biol. Chem. 289, 25262–25275 (2014).
Boguth, C. A., Singh, P., Huang, C. C. & Tesmer, J. J. Molecular basis for activation of G protein-coupled receptor kinases. EMBO J. 29, 3249–3259 (2010).
Noble, B., Kallal, L. A., Pausch, M. H. & Benovic, J. L. Development of a yeast bioassay to characterize G protein-coupled receptor kinases. Identification of an NH2-terminal region essential for receptor phosphorylation. J. Biol. Chem. 278, 47466–47476 (2003).
Palczewski, K., Buczyłko, J., Lebioda, L., Crabb, J. W. & Polans, A. S. Identification of the N-terminal region in rhodopsin kinase involved in its interaction with rhodopsin. J. Biol. Chem. 268, 6004–6013 (1993).
Lefkowitz, R. J. Seven transmembrane receptors: something old, something new. Acta Physiol. (Oxf.) 190, 9–19 (2007).
Brinks, H. & Koch, W. J. Targeting G protein-coupled receptor kinases (GRKs) in heart failure. Drug Discov. Today Dis. Mech. 7, e129–e134 (2010).
Nogués, L. et al. G protein-coupled receptor kinases (GRKs) in tumorigenesis and cancer progression: GPCR regulators and signaling hubs. Semin. Cancer Biol. 48, 78–90 (2018).
Kannan, N., Haste, N., Taylor, S. S. & Neuwald, A. F. The hallmark of AGC kinase functional divergence is its C-terminal tail, a cis-acting regulatory module. Proc. Natl Acad. Sci. USA 104, 1272–1277 (2007).
Lodowski, D. T., Pitcher, J. A., Capel, W. D., Lefkowitz, R. J. & Tesmer, J. J. Keeping G proteins at bay: a complex between G protein-coupled receptor kinase 2 and Gβγ. Science 300, 1256–1262 (2003).
Gao, Y. et al. Structures of the rhodopsin-transducin complex: insights into G-protein activation. Mol. Cell 75, 781–790.e3 (2019).
Maeda, S., Qu, Q., Robertson, M. J., Skiniotis, G. & Kobilka, B. K. Structures of the M1 and M2 muscarinic acetylcholine receptor/G-protein complexes. Science 364, 552–557 (2019).
Huang, W. et al. Structure of the neurotensin receptor 1 in complex with β-arrestin 1. Nature 579, 303–308 (2020).
Staus, D. P. et al. Structure of the M2 muscarinic receptor–β-arrestin complex in a lipid nanodisc. Nature 579, 297–302 (2020).
Kato, H. E. et al. Conformational transitions of a neurotensin receptor 1–Gi1 complex. Nature 572, 80–85 (2019).
Pulvermüller, A., Palczewski, K. & Hofmann, K. P. Interaction between photoactivated rhodopsin and its kinase: stability and kinetics of complex formation. Biochemistry 32, 14082–14088 (1993).
Kühn, H. & Dreyer, W. J. Light dependent phosphorylation of rhodopsin by ATP. FEBS Lett. 20, 1–6 (1972).
Kühn, H., Cook, J. H. & Dreyer, W. J. Phosphorylation of rhodopsin in bovine photoreceptor membranes. A dark reaction after illumination. Biochemistry 12, 2495–2502 (1973).
Clifford-Nunn, B., Showalter, H. D. & Andrews, P. C. Quaternary diamines as mass spectrometry cleavable crosslinkers for protein interactions. J. Am. Soc. Mass Spectrom. 23, 201–212 (2012).
Hagen, S. E. et al. Synthesis of CID-cleavable protein crosslinking agents containing quaternary amines for structural mass spectrometry. Org. Biomol. Chem. 16, 8245–8248 (2018).
Bayburt, T. H. et al. Monomeric rhodopsin is sufficient for normal rhodopsin kinase (GRK1) phosphorylation and arrestin-1 binding. J. Biol. Chem. 286, 1420–1428 (2011).
Palczewski, K., Kahn, N. & Hargrave, P. A. Nucleoside inhibitors of rhodopsin kinase. Biochemistry 29, 6276–6282 (1990).
Waldschmidt, H. V. et al. Structure-based design, synthesis, and biological evaluation of highly selective and potent G protein-coupled receptor kinase 2 inhibitors. J. Med. Chem. 59, 3793–3807 (2016).
Komolov, K. E. et al. Structure of a GRK5–calmodulin complex reveals molecular mechanism of GRK activation and substrate targeting. Mol. Cell 81, 323–339.e11 (2021).
Huang, C. C., Yoshino-Koh, K. & Tesmer, J. J. G. A surface of the kinase domain critical for the allosteric activation of G protein-coupled receptor kinases. J. Biol. Chem. 284, 17206–17215 (2009).
Madhusudan, A., Akamine, P., Xuong, N. H. & Taylor, S. S. Crystal structure of a transition state mimic of the catalytic subunit of cAMP-dependent protein kinase. Nat. Struct. Biol. 9, 273–277 (2002).
Yao, X. Q. et al. Navigating the conformational landscape of G protein-coupled receptor kinases during allosteric activation. J. Biol. Chem. 292, 16032–16043 (2017).
Zhou, X. E. et al. Identification of phosphorylation codes for arrestin recruitment by G protein-coupled receptors. Cell 170, 457–469.e13 (2017).
Farrens, D. L., Altenbach, C., Yang, K., Hubbell, W. L. & Khorana, H. G. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science 274, 768–770 (1996).
Pao, C. S., Barker, B. L. & Benovic, J. L. Role of the amino terminus of G protein-coupled receptor kinase 2 in receptor phosphorylation. Biochemistry 48, 7325–7333 (2009).
Huang, C. C., Orban, T., Jastrzebska, B., Palczewski, K. & Tesmer, J. J. Activation of G protein-coupled receptor kinase 1 involves interactions between its N-terminal region and its kinase domain. Biochemistry 50, 1940–1949 (2011).
Jones Brunette, A. M., Sinha, A., David, L. & Farrens, D. L. Evidence that the rhodopsin kinase (GRK1) N-terminus and the transducin Gα C-terminus interact with the same “hydrophobic patch” on rhodopsin TM5. Biochemistry 55, 3123–3135 (2016).
Kelleher, D. J. & Johnson, G. L. Characterization of rhodopsin kinase purified from bovine rod outer segments. J. Biol. Chem. 265, 2632–2639 (1990).
Buczyłko, J., Gutmann, C. & Palczewski, K. Regulation of rhodopsin kinase by autophosphorylation. Proc. Natl Acad. Sci. USA 88, 2568–2572 (1991).
Kunapuli, P., Gurevich, V. V. & Benovic, J. L. Phospholipid-stimulated autophosphorylation activates the G protein-coupled receptor kinase GRK5. J. Biol. Chem. 269, 10209–10212 (1994).
Premont, R. T., Koch, W. J., Inglese, J. & Lefkowitz, R. J. Identification, purification, and characterization of GRK5, a member of the family of G protein-coupled receptor kinases. J. Biol. Chem. 269, 6832–6841 (1994).
Ping, Y. Q. et al. Structures of the glucocorticoid-bound adhesion receptor GPR97-Go complex. Nature 589, 620–626 (2021).
Dhami, G. K. & Ferguson, S. S. Regulation of metabotropic glutamate receptor signaling, desensitization and endocytosis. Pharmacol. Ther. 111, 260–271 (2006).
Dhami, G. K. et al. G Protein-coupled receptor kinase 2 regulator of G protein signaling homology domain binds to both metabotropic glutamate receptor 1a and Galphaq to attenuate signaling. J. Biol. Chem. 279, 16614–16620 (2004).
Iacovelli, L. et al. Regulation of group II metabotropic glutamate receptors by G protein-coupled receptor kinases: mGlu2 receptors are resistant to homologous desensitization. Mol. Pharmacol. 75, 991–1003 (2009).
Ohguro, H., Palczewski, K., Ericsson, L. H., Walsh, K. A. & Johnson, R. S. Sequential phosphorylation of rhodopsin at multiple sites. Biochemistry 32, 5718–5724 (1993).
Yang, J. et al. Crystal structure of an activated Akt/protein kinase B ternary complex with GSK3-peptide and AMP-PNP. Nat. Struct. Biol. 9, 940–944 (2002).
Reiter, E., Ahn, S., Shukla, A. K. & Lefkowitz, R. J. Molecular mechanism of β-arrestin-biased agonism at seven-transmembrane receptors. Annu. Rev. Pharmacol. Toxicol. 52, 179–197 (2012).
Nobles, K. N. et al. Distinct phosphorylation sites on the β(2)-adrenergic receptor establish a barcode that encodes differential functions of β-arrestin. Sci. Signal. 4, ra51 (2011).
Shoemaker, B. A., Portman, J. J. & Wolynes, P. G. Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc. Natl Acad. Sci. USA 97, 8868–8873 (2000).
Singh, P., Wang, B., Maeda, T., Palczewski, K. & Tesmer, J. J. Structures of rhodopsin kinase in different ligand states reveal key elements involved in G protein-coupled receptor kinase activation. J. Biol. Chem. 283, 14053–14062 (2008).
Palczewski, K. et al. Crystal structure of rhodopsin: a G protein-coupled receptor. Science 289, 739–745 (2000).
Pitcher, J. A. et al. Feedback inhibition of G protein-coupled receptor kinase 2 (GRK2) activity by extracellular signal-regulated kinases. J. Biol. Chem. 274, 34531–34534 (1999).
Yu, Q. M. et al. The amino terminus with a conserved glutamic acid of G protein-coupled receptor kinases is indispensable for their ability to phosphorylate photoactivated rhodopsin. J. Neurochem. 73, 1222–1227 (1999).
Sterne-Marr, R. et al. GRK2 activation by receptors: role of the kinase large lobe and carboxyl-terminal tail. Biochemistry 48, 4285–4293 (2009).
Komolov, K. E., Bhardwaj, A. & Benovic, J. L. Atomic structure of GRK5 reveals distinct structural features novel for G protein-coupled receptor kinases. J. Biol. Chem. 290, 20629–20647 (2015).
Lodowski, D. T. et al. The role of Gβγ and domain interfaces in the activation of G protein-coupled receptor kinase 2. Biochemistry 44, 6958–6970 (2005).
Baameur, F. et al. Role for the regulator of G-protein signaling homology domain of G protein-coupled receptor kinases 5 and 6 in β2-adrenergic receptor and rhodopsin phosphorylation. Mol. Pharmacol. 77, 405–415 (2010).
Lodowski, D. T., Tesmer, V. M., Benovic, J. L. & Tesmer, J. J. The structure of G protein-coupled receptor kinase (GRK)-6 defines a second lineage of GRKs. J. Biol. Chem. 281, 16785–16793 (2006).
Papermaster, D. S. Preparation of retinal rod outer segments. Methods Enzymol. 81, 48–52 (1982).
Paduch, M. et al. Generating conformation-specific synthetic antibodies to trap proteins in selected functional states. Methods 60, 3–14 (2013).
Miller, K. R. et al. T cell receptor-like recognition of tumor in vivo by synthetic antibody fragment. PLoS ONE 7, e43746 (2012).
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Tan, Y. Z. et al. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods 14, 793–796 (2017).
Bouley, R. et al. Structural determinants influencing the potency and selectivity of indazole-paroxetine hybrid G protein-coupled receptor kinase 2 inhibitors. Mol. Pharmacol. 92, 707–717 (2017).
Ludtke, S. J., Baldwin, P. R. & Chiu, W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 (1999).
Yang, Z., Fang, J., Chittuluru, J., Asturias, F. J. & Penczek, P. A. Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure 20, 237–247 (2012).
Perez-Riverol, Y. et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019).
Rasmussen, S. G. et al. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature 477, 549–555 (2011).
Bujacz, A. Structures of bovine, equine and leporine serum albumin. Acta Crystallogr. D 68, 1278–1289 (2012).
Combe, C. W., Fischer, L. & Rappsilber, J. xiNET: cross-link network maps with residue resolution. Mol. Cell. Proteomics 14, 1137–1147 (2015).
Acknowledgements
This work was supported by National Institutes of Health grants HL071818, HL122416, and CA221289 (to J.J.G.T.), GM117372 (to A.A.K.), GM095832, GM105942, GM109896, and GM105920 (to P.C.A.), an American Heart Association Post-Doctoral Fellowship (19POST34450193, to Q.C.), and National Science Foundation grant MCB-1517617 (to X.-Q.Y.). J.J.G.T. was also supported by the Walther Cancer Foundation. We thank the Purdue Cryo-EM Facility for equipment access and support.
Author information
Authors and Affiliations
Contributions
Q.C. and J.J.G.T. conceptualized the study. Q.C. produced and purified rhodopsin and GRK1, and performed crosslinking and kinetic assays. S.M., Q.C., J.J.G.T., and A.A.K. selected the Fabs. Q.C., J.J.G.T., Z.L., and L.C. collected data and performed structure determinations of Rho*−GRK1 and Rho*−GRK1(S5E/EE). Q.C. and J.J.G.T collected data and performed structure determinations of Rho*−GRK1(S5E/EE)−Fab1 and Rho*−GRK1(S5E/EE)−Fab6. T.K. assisted with cryo-EM data collection for all four reconstructions. M.P. and P.C.A. performed the mass spectrometry analysis. D.P. collected and performed negative-stain EM analysis. C.-L.C., J.J.G.T and Q.C. performed docking and molecular simulations. X.-Q.Y. performed the PCA analysis. Q.C. wrote the original draft and all authors further edited the manuscript. Q.C., J.J.G.T. and P.C.A. contributed funding.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Vsevolod Gurevich, Carol Robinson and Patrick M. Sexton for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Modulation of GRK1 activity by anionic lipids, Rho* C-terminal modifications, and Fab fragments.
a, Kinetic analysis of GRK1 phosphorylation of Rho* in ROS or LMNG or LMNG + c8-PtdIns(4,5)P2. Mean ± s.d., n = 4 technical replicates. Reactions were performed in 50 mM HEPES (pH 8.0), 10 mM MgCl2 for 2 min at room temperature. b, Kinetic analysis of GRK1 phosphorylation of Rho* in ROS or POPC nanodiscs containing 40% POPG or 40% POPS or 10% PtdIns(4,5)P2. Mean ± s.d., n = 3 technical replicates. Reactions were performed in 50 mM HEPES (pH 8.0), 10 mM MgCl2 for 2 min at room temperature. c, Representative Michaelis–Menten kinetics measurement with varying ATP. Rho* was reconstituted with POPC nanodiscs containing 40% POPG (black squares) or 40% POPS (black triangles) and compared with Rho* in ROS (black circles). GRK1(5A) activity was greatly diminished relative to GRK1 under these conditions (blue symbols) but was still somewhat responsive to anionic lipids. d, Approximate location of the five positively charged residues places them close to the lipid bilayer. e, The crosslinking yield (corresponding to the amount of complex formed divided by the sum of the input GRK1 and Rho*) of GRK1(5A) was significantly lower than that of GRK1 in the presence of c8-PtdIns(4,5)P2 but not in its absence. The crosslinking level of GRK1(5A) was compared with that of GRK1 using a two-sided t-test (n = 3 technical replicates) in the presence or absence of c8-PtdIns(4,5)P2. For gel source data, see Supplementary Fig. 5f. f, The crosslinking yield of p-Rho* (with estimated 6–8 phosphates out of 7 incorporated based on a standard curve) with GRK1 was compared with that of unphosphorylated rhodopsin using two-sided t-test (n = 4 technical replicates). For gel source data, see Supplementary Fig. 5h. g, The crosslinking yield of Asp-N-truncated Rho* (cleavage site N-terminal to Asp329) was compared with that of full-length Rho* using two-sided t-test (n = 5 technical replicates; N.S., not significant). For gel source data, see Supplementary Fig. 5i. The GRK1(K479R) mutant, which eliminates the other prominent crosslinking site, also did not affect our ability to trap the complex (data not shown). Mean ± s.d. h, ELISA analysis of Fab1 and Fab6 binding to GRK1 yielded EC50 values of 2 and 5 nM, respectively (n = 3 technical replicates). Mean ± s.d. i, Michaelis–Menten analysis of GRK1 in the absence or presence of threefold molar excess Fab1 or Fab6 (n = 3 technical replicates). Data were normalized to the fit Vmax, Rho of GRK1. Mean ± s.d.
Extended Data Fig. 2 Workflow of cryo-EM data processing and resolution analysis of Rho*−GRK1.
a, Representative raw cryo-EM micrograph from a total of 2,542. From these, 2.7 million particles were automatically picked in RELION-3 and were used to generate 2D class averages (shown are representative good classes). After screening out bad classes, 705,966 particles remained. Three major classes from 3D classification were generated in RELION-3 using an initial model generated by cryoSPARC. Class 2 (183,717 particles) showed the best quality and was selected for 3D auto-refinement, resulting in a final map at a global resolution of 7.0 Å (FSC cut-off = 0.143). b, Directional FSC indicated that the resolutions in the x and z directions are similar to the global resolution, whereas the resolution in the y direction is lower because fewer particles have this axis resolved. c, Plots of the global FSC together with the spread of directional resolution values defined by ±1σ from the mean and a histogram of 100 such values evenly sampled over the 3D FSC. d, Local resolution map as estimated by RELION-3.
Extended Data Fig. 3 Workflow of cryo-EM data processing and resolution analysis of Rho*−GRK1(S5E/EE).
a, Representative raw cryo-EM micrograph from a total of 2,130. From these, 2.7 million particles were automatically picked in cryoSPARC and were used to generate 2D class averages (shown are representative good classes). After screening out bad classes, the 584,674 remaining particles were imported to RELION-3 and used to generate six major classes from 3D classification in RELION-3 using an initial model generated by cryoSPARC. Class 6 (132,721 particles) showed the best quality and was selected for 3D auto-refinement, resulting in a final map at a global resolution of 5.8 Å (FSC cut-off = 0.143). b, Directional FSC indicates that the resolutions in the x and z directions are similar to the global resolution, whereas the resolution in the y direction is lower. c, Plots of the global FSC together with the spread of directional resolution values defined by ±1σ from the mean and a histogram of 100 such values evenly sampled over the 3D FSC. d, Local resolution map as estimated by RELION-3.
Extended Data Fig. 4 Workflow of cryo-EM data processing and resolution analysis of Rho*−GRK1(S5E/EE)−Fab1.
a, Representative raw cryo-EM micrograph from a total of 5,501. From these, 5.7 million particles were automatically picked in cryoSPARC and were used to generate 2D class averages (shown are representative good classes). After screening out bad classes, the 421,682 remaining particles were further processed using heterogeneous refinement. Classes 2 and 3 showed similar quality and were selected for homogeneous refinement and then non-uniform refinement in cryoSPARC, resulting in a final map at a global resolution of 4.1 Å (FSC cut-off = 0.143). b, Directional FSC indicates that the resolutions in the x and z directions are similar to the global resolution, whereas the resolution in the y direction is lower. c, Plots of the global FSC together with the spread of directional resolution values defined by ±1σ from the mean and a histogram of 100 such values evenly sampled over the 3D FSC. d, Local resolution map as estimated by cryoSPARC.
Extended Data Fig. 5 Flow chart of cryo-EM data processing and resolution analysis of Rho*−GRK1(S5E/EE)−Fab6, and comparison of the Fab1 and Fab6 complexes.
a, A representative raw cryo-EM micrograph from a total of 8,358. From these, 7.5 million particles were automatically picked in cryoSPARC and were used to generate 2D class averages (shown are representative good classes). After screening out bad classes, the 352,864 remaining particles were further processed using heterogeneous refinement. Classes 2 and 3 showed similar quality and were selected for homogeneous refinement and then non-uniform refinement in cryoSPARC. The resolution of the final map is estimated to be 4.0 Å. Global resolution was determined by FSC with a cut-off of 0.143. b, Directional FSC indicates that the resolutions in the x and z directions are similar to the globular resolution, whereas the resolution in the y direction is lower. c, Plots of the global FSC together with the spread of directional resolution values defined by ±1σ from the mean and a histogram of 100 such values evenly sampled over the 3D FSC. d, Local resolution map as estimated by cryoSPARC. e, Overlay of Rho* from the Fab1 (red) and Fab6 (blue) complexes after alignment of the small lobes of their kinase domains. Rho* pivots by about 5.5°, largely around an axis roughly parallel to a vector joining the ICL1 and ICL3 loops of the receptor. The key interactions between Rho* and GRK1, namely that of TM3, TM5 and TM6 with αN, and ICL1 with the C-terminal end of the AST, stay intact. This is because the αN helix and the N-terminal half of the AST loop region of GRK1 shift with the receptor, which is reasonable given that these elements are known to be dynamic from crystal structures of GRKs. Motion of these elements is likely to be responsible for the observed difference in kinase domain conformation exhibited by the Fab1 and Fab6 complexes because they directly affect the hinge region of the kinase domain. There are, however, small conformational differences at the end of TM1 and the beginning of the ICL1 loop in Rho* that probably occur because the bound GPCR is trying to maintain optimal interactions with phosphosites at the end of the AST in each state. The conformation of ICL2 is also likely to change between the structures, but this region could not be accurately modelled. Because our PC analysis (Fig. 2e) suggests that the Fab6 complex features a less active conformation of the GRK1 kinase domain, it may represent an intermediate state in which αN and AST have engaged the receptor, but the kinase domain has yet to fully adopt a transition state-like conformation.
Extended Data Fig. 6 Assessment of ligand density, of the presence of all-trans retinal, and of the conformational heterogeneity of the GRK1 RH domain.
a, Electron density of all-trans retinal, contoured at 17σ (Fab1 map) and 13σ (Fab6 map); Sgv, contoured at 24σ (Fab1 map) and 26σ (Fab6 map); and αN, contoured at 21σ (Fab1 map) and 25σ (Fab6 map). b, Light- and GRK ligand-dependence of the crosslinking reaction between rhodopsin and GRK1. 11-cis retinal undergoes isomerization upon light exposure to all-trans retinal, which serves as a full agonist for rhodopsin. The crosslinking level of GRK1 with rhodopsin in the dark (n = 4 technical replicates) and in the light with excess 11-cis retinal (n = 3 technical replicates) were compared to that of GRK1 with rhodopsin in the light using one-way ANOVA followed by a Dunnett’s multiple comparisons test. Mean ± s.d. c, Density for the RH domain was not observed by cryo-EM in any of our reconstructions. The map of Rho*−GRK1(S5E/EE)−Fab1 is shown here as an example. d, A representative negative-stain EM micrograph of the Rho*−GRK1 complex solubilized in LMNG (left) along with representative 2D averages (right), indicating heterogeneity in the bound GRK1. The smaller, variably positioned domain is interpreted as the RH domain.
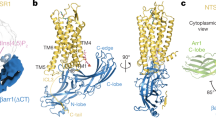
Extended Data Fig. 7 GRK αN interactions, kinase domain conformational changes, and comparison of the interactions between Rho* and its three principal downstream targets.
a–d, The GRK N terminus folds into a helix (αN) that packs against the small lobe and AST of the kinase domain, forming a docking side for Rho* (a, b; GRK1 with Fab1 and Fab6 complexes, respectively), Ca2+·CaM (c; GRK5 in PDB entry 6PJX27), or a twofold related crystal lattice contact (d; GRK6 in PDB entry 3NYN6). The interfaces shown are mediated by the same highly conserved hydrophobic residues (side chains shown with spheres). e, The cytoplasmic cleft and ICL1 of activated β2AR in its G-protein-bound conformation (PDB entry 3SN671) readily accommodates αN and AST of GRK1. f–h, Kinase domain small lobes from different ‘active’ GRK structures were aligned to highlight differences in closure. i–l, Comparison of downstream proteins bound to the cytoplasmic surface of Rho*. The side chains of residues interacting with GRK1 in the Fab1 complex (i; PDB entry 7MTA), GRK1 in the Fab6 complex (j; PDB entry 7MTB), Gαt (transducin) (k; PDB entry 6OYA14) and arrestin-1 (l; PDB entry 5W0P31) are shown as yellow spheres. Note that αN of GRK1 and α5 of Gα bind with opposite polarity. m, n, Cartoon representations of GRK1 αN helix docked to Rho* in the Fab1 (m) and Fab6 (n) models. o, Gαt C terminus bound to Rho* (PDB entry 6OYA14). p, Arrestin-1 finger loop bound to Rho* (PDB entry 5W0P31).
Extended Data Fig. 8 Interactions of GRK1 with intracellular loops of Rho* and development of autophosphorylation mimetic variants.
a, Interactions of GRK1 αN with the cytoplasmic cleft and H8 of Rho*. GRK1-Ser5 was modelled in a phosphorylated state to demonstrate proximity to Rho*-Lys311 and Arg314. b, GRK1 AST interaction with ICL1. The key participating residues are shown with stick side chains. Ser488 and Thr489 are autophosphorylation sites in GRK1. c–g, Kinetic and crosslinking analysis of GRK1 and phosphomimetic mutants of Rho* autophosphorylation (S5E; S488E and T489E (EE); S488D and T489D (DD); S5E, S488E, and T489E (S5E/EE)). One-way ANOVA followed by Dunnett’s multiple comparison test was carried out to compare each mutant with GRK1. Reactions were performed in 50 mM HEPES (pH 8.0), 10 mM MgCl2 for 2 min at room temperature. Data were normalized to the Vmax,Rho or Vmax,ATP of GRK1. For Rho kinetics: S5E, n = 4; EE, n = 5; DD, n = 3; S5E/EE, n = 3. For ATP kinetics: S5E, n = 3; EE, n = 3; DD, n = 4; S5E/EE, n = 4 (all technical replicates). All data shown as mean ± s.d. f, Time courses of tubulin phosphorylation by GRK1 and variants are similar. Data were normalized to the phosphorylation level of tubulin by GRK1 at 30 min (n = 3 technical replicates). g, Crosslinking yield of GRK1 variants relative to GRK1 (n = 3 technical replicates). For gel source data, see Supplementary Fig. 5. h, Interaction of the GRK1 small lobe with ICL2 of Rho*. The GRK1 α0 helix from a basal ATP-bound structure (PDB entry 3C4Z49) is modelled by aligning its small lobe with that in the Rho*−GRK1(S5E/EE)−Fab1 complex, demonstrating a potential clash between ICL2 and α0. i, Interaction of GRK1 AST with ICL3.
Extended Data Fig. 9 MC4 crosslinker properties and liquid chromatography tandem mass spectrometry peptide coverage of the Rho*−GRK1 complex.
a, MC4 has two symmetrical amine-reactive NHS esters separated by a C4 linker attached to a quaternary N-morpholine group. *Conformational analyses of in vacuo MD simulations suggest an average carboxy C–C distance of approximately 6 Å. b, Characterization of MC4 reactive distance using BSA. The y-axis shows the frequency (expressed as %) of experimentally identified crosslinked lysine pairs within a certain Cα–Cα distance (x-axis) based on a BSA crystal structure (PDB entry 4F5S72). c, Peptide sequence coverage obtained for the purified crosslinked Rho*−GRK1 complex after multistep digestion with Staphylococcus aureus Protease V8 (GluC)/mix (1:1 mixture of GluC/trypsin and GluC/chymotrypsin) or GluC/trypsin. d, Intramolecular and intermolecular crosslinks summarized from Supplementary Data Tables 2, 3 are shown with a bar plot generated by XiNET73. Dashed green lines represent rare intermolecular crosslinks that were too distant to react in our cryo-EM models.
Supplementary information
Supplementary Information
This file contains Supplementary Methods, Supplementary Figures 1-5, Supplementary Tables 1-4, a Supplementary Discussion and Supplementary References.
Video 1
: 360° rotation of the model for the Rho*−GRK1S5E/EE−Fab1 complex fitted into its cryo-EM map. Corresponding map was contoured at 6 σ with the detergent micelle density omitted for clarity. Videos were generated using UCSF Chimera.
Video 2
: 360° rotation of the model for the Rho*−GRK1S5E/EE−Fab6 complex fitted into its cryo-EM map Corresponding map was contoured at 6 σ with the detergent micelle density omitted for clarity. Videos were generated using UCSF Chimera.
Video 3
: Conformational change associated with principal component 1 (PC1) derived from known GRK and PKA kinase domain structures PC1 represents the dominant conformational change observed in the kinase domain (10% of variance). Images were generated by PyMOL2 (Schrödinger LLC) and videos were rendered with Photoshop 2020 (Adobe Inc).
Video 4
: Conformational changes associated with principal component 2 (PC2) derived from known GRK and PKA kinase domain structures PC2 represents the second largest conformational change observed in the kinase domain (10% of variance). Images were generated by PyMOL2 (Schrödinger LLC) and videos were rendered with Photoshop 2020 (Adobe Inc).
Rights and permissions
About this article
Cite this article
Chen, Q., Plasencia, M., Li, Z. et al. Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1. Nature 595, 600–605 (2021). https://doi.org/10.1038/s41586-021-03721-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03721-x
This article is cited by
-
Cryo-electron microscopy for GPCR research and drug discovery in endocrinology and metabolism
Nature Reviews Endocrinology (2024)
-
GPR161 structure uncovers the redundant role of sterol-regulated ciliary cAMP signaling in the Hedgehog pathway
Nature Structural & Molecular Biology (2024)
-
Structure, function and drug discovery of GPCR signaling
Molecular Biomedicine (2023)
-
GPCR activation and GRK2 assembly by a biased intracellular agonist
Nature (2023)
-
Fabrication of helix–fiber composites with mechanically coupled core-wrapping for programmable properties
Communications Materials (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.