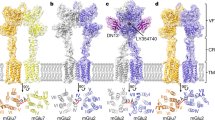
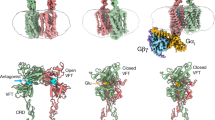
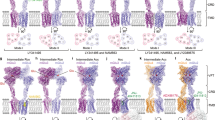
Abstract
The metabotropic glutamate receptors (mGlus) have key roles in modulating cell excitability and synaptic transmission in response to glutamate (the main excitatory neurotransmitter in the central nervous system)1. It has previously been suggested that only one receptor subunit within an mGlu homodimer is responsible for coupling to G protein during receptor activation2. However, the molecular mechanism that underlies the asymmetric signalling of mGlus remains unknown. Here we report two cryo-electron microscopy structures of human mGlu2 and mGlu4 bound to heterotrimeric Gi protein. The structures reveal a G-protein-binding site formed by three intracellular loops and helices III and IV that is distinct from the corresponding binding site in all of the other G-protein-coupled receptor (GPCR) structures. Furthermore, we observed an asymmetric dimer interface of the transmembrane domain of the receptor in the two mGlu–Gi structures. We confirmed that the asymmetric dimerization is crucial for receptor activation, which was supported by functional data; this dimerization may provide a molecular basis for the asymmetric signal transduction of mGlus. These findings offer insights into receptor signalling of class C GPCRs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Atomic coordinates and cryo-EM density maps for the structures of mGlu2–Gi1 and mGlu4–Gi3 complexes have been deposited in the PDB under identification codes 7E9G and 7E9H, respectively, and in the Electron Microscopy Data Bank under accession codes EMD-31031 and EMD-31032, respectively. The uncropped gels shown in Extended Data Figs. 1b, i, 5u are displayed in Supplementary Fig. 1. The database used in this study includes PDB 3SN6, 4OR2, 4OO9, 4XAQ, 4XAS, 5CNI, 5CNJ, 6DDE, 6LMK, 6LML, 6N51, 6N52 and 6OT0.
References
Niswender, C. M. & Conn, P. J. Metabotropic glutamate receptors: physiology, pharmacology, and disease. Annu. Rev. Pharmacol. Toxicol. 50, 295–322 (2010).
Hlavackova, V. et al. Evidence for a single heptahelical domain being turned on upon activation of a dimeric GPCR. EMBO J. 24, 499–509 (2005).
Kniazeff, J. et al. Closed state of both binding domains of homodimeric mGlu receptors is required for full activity. Nat. Struct. Mol. Biol. 11, 706–713 (2004).
Tateyama, M., Abe, H., Nakata, H., Saito, O. & Kubo, Y. Ligand-induced rearrangement of the dimeric metabotropic glutamate receptor 1α. Nat. Struct. Mol. Biol. 11, 637–642 (2004).
Matsushita, S., Nakata, H., Kubo, Y. & Tateyama, M. Ligand-induced rearrangements of the GABAB receptor revealed by fluorescence resonance energy transfer. J. Biol. Chem. 285, 10291–10299 (2010).
Rasmussen, S. G. et al. Crystal structure of the β2 adrenergic receptor–Gs protein complex. Nature 477, 549–555 (2011).
Zhang, Y. et al. Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein. Nature 546, 248–253 (2017).
Monn, J. A. et al. Design, synthesis, and pharmacological characterization of (+)-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylic acid (LY354740): a potent, selective, and orally active group 2 metabotropic glutamate receptor agonist possessing anticonvulsant and anxiolytic properties. J. Med. Chem. 40, 528–537 (1997).
Kang, H. J. et al. Selectivity and evolutionary divergence of metabotropic glutamate receptors for endogenous ligands and G proteins coupled to phospholipase C or TRP channels. J. Biol. Chem. 289, 29961–29974 (2014).
Cid, J. M. et al. Discovery of 1-butyl-3-chloro-4-(4-phenyl-1-piperidinyl)-(1H)-pyridone (JNJ-40411813): a novel positive allosteric modulator of the metabotropic glutamate 2 receptor. J. Med. Chem. 57, 6495–6512 (2014).
Jones, C. K. et al. The metabotropic glutamate receptor 4-positive allosteric modulator VU0364770 produces efficacy alone and in combination with l-DOPA or an adenosine 2A antagonist in preclinical rodent models of Parkinson’s disease. J. Pharmacol. Exp. Ther. 340, 404–421 (2012).
Scholler, P. et al. Allosteric nanobodies uncover a role of hippocampal mGlu2 receptor homodimers in contextual fear consolidation. Nat. Commun. 8, 1967 (2017).
Koehl, A. et al. Structural insights into the activation of metabotropic glutamate receptors. Nature 566, 79–84 (2019).
Monn, J. A. et al. Synthesis and pharmacological characterization of C4-disubstituted analogs of 1S,2S,5R,6S-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylate: identification of a potent, selective metabotropic glutamate receptor agonist and determination of agonist-bound human mGlu2 and mGlu3 amino terminal domain structures. J. Med. Chem. 58, 1776–1794 (2015).
Monn, J. A. et al. Synthesis and pharmacological characterization of C4-(thiotriazolyl)-substituted-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylates. Identification of (1R,2S,4R,5R,6R)-2-amino-4-(1H-1,2,4-triazol-3-ylsulfanyl)bicyclo[3.1.0]hexane-2,6-dicarboxylic acid (LY2812223), a highly potent, functionally selective mGlu2 receptor agonist. J. Med. Chem. 58, 7526–7548 (2015).
Du, J. et al. Structures of human mGlu2 and mGlu7 homo- and heterodimers. Nature https://doi.org/10.1038/s41586-021-03641-w (2021).
Shaye, H. et al. Structural basis of the activation of a metabotropic GABA receptor. Nature 584, 298–303 (2020).
Mao, C. et al. Cryo-EM structures of inactive and active GABAB receptor. Cell Res. 30, 564–573 (2020).
Francesconi, A. & Duvoisin, R. M. Role of the second and third intracellular loops of metabotropic glutamate receptors in mediating dual signal transduction activation. J. Biol. Chem. 273, 5615–5624 (1998).
Chang, W., Chen, T. H., Pratt, S. & Shoback, D. Amino acids in the second and third intracellular loops of the parathyroid Ca2+-sensing receptor mediate efficient coupling to phospholipase C. J. Biol. Chem. 275, 19955–19963 (2000).
Dixon, A. S. et al. NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem. Biol. 11, 400–408 (2016).
Qiao, A. et al. Structural basis of Gs and Gi recognition by the human glucagon receptor. Science 367, 1346–1352 (2020).
Flock, T. et al. Universal allosteric mechanism for Gα activation by GPCRs. Nature 524, 173–179 (2015).
Pin, J. P., Galvez, T. & Prézeau, L. Evolution, structure, and activation mechanism of family 3/C G-protein-coupled receptors. Pharmacol. Ther. 98, 325–354 (2003).
Lundström, L. et al. Pharmacological and molecular characterization of the positive allosteric modulators of metabotropic glutamate receptor 2. Neuropharmacology 111, 253–265 (2016).
Pérez-Benito, L. et al. Molecular switches of allosteric modulation of the metabotropic glutamate 2 receptor. Structure 25, 1153–1162.e4 (2017).
Wu, H. et al. Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science 344, 58–64 (2014).
Doré, A. S. et al. Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain. Nature 511, 557–562 (2014).
Rovira, X. et al. Overlapping binding sites drive allosteric agonism and positive cooperativity in type 4 metabotropic glutamate receptors. FASEB J. 29, 116–130 (2015).
Gregory, K. J. et al. Identification of specific ligand-receptor interactions that govern binding and cooperativity of diverse modulators to a common metabotropic glutamate receptor 5 allosteric site. ACS Chem. Neurosci. 5, 282–295 (2014).
Fukuda, J. et al. Identification of a novel transmembrane domain involved in the negative modulation of mGluR1 using a newly discovered allosteric mGluR1 antagonist, 3-cyclohexyl-5-fluoro-6-methyl-7-(2-morpholin-4-ylethoxy)-4H-chromen-4-one. Neuropharmacology 57, 438–445 (2009).
Xue, L. et al. Major ligand-induced rearrangement of the heptahelical domain interface in a GPCR dimer. Nat. Chem. Biol. 11, 134–140 (2015).
Huang, S. et al. Interdomain movements in metabotropic glutamate receptor activation. Proc. Natl Acad. Sci. USA 108, 15480–15485 (2011).
Hlavackova, V. et al. Sequential inter- and intrasubunit rearrangements during activation of dimeric metabotropic glutamate receptor 1. Sci. Signal. 5, ra59 (2012).
Kang, Y. et al. Cryo-EM structure of human rhodopsin bound to an inhibitory G protein. Nature 558, 553–558 (2018).
García-Nafría, J., Nehmé, R., Edwards, P. C. & Tate, C. G. Cryo-EM structure of the serotonin 5-HT1B receptor coupled to heterotrimeric Go. Nature 558, 620–623 (2018).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Arnold, K., Bordoli, L., Kopp, J. & Schwede, T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 (2006).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Monnier, C. et al. Trans-activation between 7TM domains: implication in heterodimeric GABAB receptor activation. EMBO J. 30, 32–42 (2011).
Kato, H. E. et al. Conformational transitions of a neurotensin receptor 1–Gi1 complex. Nature 572, 80–85 (2019).
Kingston, A. E. et al. LY341495 is a nanomolar potent and selective antagonist of group II metabotropic glutamate receptors. Neuropharmacology 37, 1–12 (1998).
Acknowledgements
The cryo-EM studies were performed at the electron microscopy facility of Shanghai Institute of Materia Medica (SIMM), Chinese Academy of Sciences. We thank Q. Wang from SIMM for cryo-EM data collection and Q. Wang from ShanghaiTech University for help with cryo-EM data processing. This work was supported by the National Science Foundation of China grants 31825010 (B.W.), 81872915 and 82073904 (M.-W.W.), and 81773792 and 81973373 (D.Y.), National Key R&D Program of China 2018YFA0507000 (B.W., Q.Z. and M.-W.W.), CAS Strategic Priority Research Program XDB37030100 (B.W. and Q.Z.) and National Science & Technology Major Project of China – Key New Drug Creation and Manufacturing Program 2018ZX09735–001 (M.-W.W.) and 2018ZX09711002–002–005 (D.Y.).
Author information
Authors and Affiliations
Contributions
S.L. developed the protein expression and purification procedures, prepared the protein samples for cryo-EM, performed signalling assays and helped with manuscript preparation. S.H. prepared cryo-samples, collected cryo-EM data and performed cryo-EM data processing and analysis, model building and structure refinement. X. Cai, A.D., Yan Zhou and Y.C. developed and performed NanoBiT assays. Q.T. helped with signalling assays and negative-stain electron microscopy data acquisition and analysis. K.Z. and X.W. helped with protein preparation and signalling assays. D.W. performed crosslinking assays and helped with manuscript preparation. J.D. helped with data analysis. C.Y. and X. Chu expressed the proteins. Yu Zhou and H.L. provided the mGlu2 PAM, and helped with data analysis and interpretation. J.L. helped with data analysis and interpretation, and edited the manuscript. D.Y. and M.-W.W. oversaw the NanoBiT assay, helped with data analysis and interpretation, and edited the manuscript. Q.Z. and B.W. initiated the project, planned and analysed experiments, supervised the research and wrote the manuscript with input from all co-authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Karen Gregory, Guillaume Lebon and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Sample preparation and cryo-EM processing of the mGlu2–Gi1 and mGlu4–Gi3 complexes.
a–g, Results of the mGlu2–Gi1 complex. a, Comparison of protein samples of the wild-type mGlu2 (WT) and the mGlu2 construct (including truncation of the C-terminal residues V826–L872 and introduction of mutation S601A) used to solve the mGlu2–Gi1 structure. The curves of SEC of the purified protein samples show higher protein yield of the construct (red) than the WT (black). b, Analytical SEC and SDS–PAGE (Coomassie blue stain) of the purified complex. Four independent experiments were performed with similar results. Results from a representative experiment are shown. For gel source data, see Supplementary Fig. 1. c, Representative cryo-EM image from four independent experiments with similar results. d, Two-dimensional averages. e, Gold-standard Fourier shell correlation (FSC) curve for the local refinement of the TMDs and Gi1, showing a resolution of 3.5 Å. f, Gold-standard FSC curve for the local refinement of the ECDs, showing a resolution of 3.1 Å. g, Composite cryo-EM map coloured according to local resolution (in Å). h–n, Results of the mGlu4–Gi3 complex. h, Comparison of protein samples of the mGlu4–Gi1 and mGlu4–Gi3 complexes. The SEC curves of the purified protein samples show better protein homogeneity of the mGlu4–Gi3 complex (red) than the mGlu4–Gi1 complex (black). i, Analytical SEC and SDS–PAGE (Coomassie blue stain) of the purified complex. Six independent experiments were performed with similar results. Results from a representative experiment are shown. For gel source data, see Supplementary Fig. 1. j, Representative cryo-EM image from four independent experiments with similar results. k, Two-dimensional averages. l, Gold-standard FSC curve, showing an overall resolution of 4.0 Å. m, Gold-standard FSC curve for the local refinement of the ECDs, showing a resolution of 3.1 Å. n, Composite cryo-EM map coloured according to local resolution (in Å).
Extended Data Fig. 2 Cryo-EM processing workflow of mGlu2–Gi1 and mGlu4–Gi3 complexes.
a, Data processing workflow of mGlu2–Gi1. b, Angular distribution of the ECDs, and TMDs and Gi1 reconstructions, in mGlu2–Gi1. c, Data processing workflow of mGlu4–Gi3. d, Angular distribution of the ECDs, and mGlu4 and Gi3 reconstructions, in mGlu4–Gi3.
Extended Data Fig. 3 Cryo-EM density maps of the mGlu2–Gi1 and mGlu4–Gi3 structures.
a, mGlu2–Gi1. b, mGlu4–Gi3. Cryo-EM density maps and models of the two structures are shown for helices III, V and VI and ICL1, ICL2 and ICL3 in the mGluG subunit, helices I, VI and VII in the mGlufree subunit, Gαi αN helix and α5 helix, and ligands LY354740, JNJ-40411813 and L-SOP. The models are shown as sticks and coloured blue (mGlu2G), cyan (mGlu2free), orange (mGlu4G), yellow (mGlu4free), green (Gαi), magenta (JNJ-40411813), red (LY354740) and purple (L-SOP). The density maps are coloured grey.
Extended Data Fig. 4 Structural comparison with other GPCR structures and binding modes of Gi and PAM.
a, Comparison of VFT conformation. The VFT domains in the Gi-bound structures of mGlu2 and mGlu4 as well as previously published structures of agonist-bound mGlu5 (PDB code 6N51) and mGlu2 VFT bound to LY354740 (PDB code 4XAQ) are shown in cartoon representation and coloured blue, orange, pink and grey, respectively. The agonists in these structures are shown as sticks. b, Comparison of agonist binding mode. The agonists in the structures of mGlu2–Gi1, mGlu4–Gi3, mGlu5, mGlu2 VFT–LY354740, mGlu2 VFT–LY2934747 (PDB code 4XAS), mGlu2 VFT–glutamate (PDB code 5CNI) and mGlu2 VFT–LY2812223 (PDB code 5CNJ) are shown as sticks with blue, orange, pink, grey, red, green and cyan carbons, respectively. Only the receptors in the mGlu2–Gi1 and mGlu4–Gi3 structures are shown for clarity. c, Conformations of CRD and ECL2 in the mGlu2–Gi1 and mGlu4–Gi3 complexes. The two subunits in the mGlu2–Gi1 structure are coloured blue and cyan, respectively. The two subunits in the mGlu4–Gi3 structure are coloured orange and yellow, respectively. The cryo-EM maps are coloured grey. d, Comparison of the TMD helical bundles in the structures of mGlu2–Gi1 (mGlu2G), mGlu4–Gi3 (mGlu4G), inactive mGlu2 (mGlu2inactive) and inactive mGlu5 (mGlu5inactive; PDB code 6N52). e, Structural comparison of the TMDs in the mGlu2–Gi1 structure. f, Structural comparison of the TMDs in the mGlu4–Gi3 structure. g, Binding pocket for the C terminus of Gαi α5 helix in mGlu2. The receptor is shown in blue cartoon and surface representations in an intracellular view. The Gαi α5 helix is shown in green cartoon representation. h, Binding pocket for the C terminus of Gαi α5 helix in mGlu4. The receptor is shown in orange cartoon and surface representations in an intracellular view. i, A hypothetical dual-Gi-binding model of mGlu2. The TMDs of the mGlu2 dimer are shown as blue and cyan cartoon, respectively. The two Gi proteins are displayed as green and grey surface, respectively. The panel shows that the two Gi proteins form a clash if both mGlu2 subunits bind to Gi. The clash is highlighted by a red circle. j, Comparison of the positions of the C terminus of Gα α5 helix in some G-protein-bound GPCR structures. The mGlu2–Gi1 and mGlu4–Gi3 structures and the previously determined structures of β2AR–Gs, μOR–Gi, GCGR–Gs, GCGR–Gi and SMO–Gi (PDB codes 3SN6, 6DDE, 6LMK, 6LML and 6OT0, respectively) are coloured blue, orange, dark red, yellow, magenta, pink and cyan, respectively. Only the helical bundle of the receptors and Gα α5 helix are shown for clarity. The Gα α5 helices in the structures are highlighted by a red dashed box. The red arrow indicates the shift of the α5 C terminus in the Gi-bound mGlu structures relative to the other known GPCR–G protein structures. k, Comparison of the Gi binding pose in mGlu2 and a class A GPCR. The structures of mGlu2–Gi1 and μOR–Gi are shown in blue and yellow cartoon representation, respectively. The Gi proteins in the two structures are also shown as surface. The red arrow indicates the movement of the Gi protein in the mGlu2–Gi1 structure relative to the μOR–Gi structure. l, Comparison of allosteric modulator binding sites in mGlus. The receptors in the mGlu2–Gi1 structure and the previously determined structures of mGlu1 TMD–FITM and mGlu5 TMD–mavoglurant (PDB codes 4OR2 and 4OO9, respectively) are shown in cartoon representation and coloured blue, dark red and light gold, respectively. The allosteric modulators JNJ-40411813, FITM and mavoglurant are shown as sticks and coloured magenta, dark red and light gold, respectively. m, n, Comparison of the cryo-EM maps of the PAM-binding site in the mGlu2G (m) and mGlu2free (n) subunits. The mGlu2G and mGlu2free subunits are coloured blue and cyan, respectively. The PAM JNJ-40411813 is shown as magenta sticks. The residues within the PAM-binding pocket are shown as sticks. The cryo-EM maps are coloured grey. o, Comparison of binding modes between the PAM and NAM in mGlu2. The receptors in the structures of mGlu2–Gi1 and mGlu2 TMD–NAM56316 are coloured blue and grey, respectively. The PAM JNJ-40411813 and NAM NAM563 are shown as magenta and yellow sticks, respectively. The residue W7736.50 that forms a close contact with the PAM and NAM in the structures is shown as sticks. The red arrows indicate the rotamer conformational change of W7736.50 and downward shift of helix VI in the mGlu2G subunit of the mGlu2–Gi1 complex relative to the NAM-bound inactive structure. p, Comparison of the mGlu2–Gi1 and mGlu2 TMD–NAM563 structures. The two subunits in the mGlu2–Gi1 structure are coloured blue and cyan, respectively. The mGlu2 TMD–NAM563 structure16 is in grey. The residues I7716.48 in the mGlu2G subunit and L8007.38 and V8047.42 in the mGlu2free subunit that form contacts are shown as sticks. The residue I7716.48 in the mGlu2 TMD–NAM563 structure is also shown as sticks. The red arrow indicates the movement of helix VI in the Gi-bound structure relative to the inactive structure.
Extended Data Fig. 5 Gi activation, signalling and crosslinking assays of mGlu2 and mGlu4.
a, e, i, LY354740-induced IP accumulation assay of wild-type mGlu2 (WT) and mutants of the mGlu2 residues involved in Gi binding. b, f, j, LY354740-induced Gi activation assay of WT mGlu2 and mutants of the mGlu2 residues involved in Gi binding using NanoBiT. c, g, k, Glutamate- and VU0364770-induced IP accumulation assay of WT mGlu4 and mutants of the mGlu4 residues involved in Gi binding. d, h, l, Glutamate- and VU0364770-induced Gi activation assay of WT mGlu4 and mutants of the mGlu4 residues involved in Gi binding using NanoBiT. m, q, JNJ-40411813-induced IP accumulation assay of WT mGlu2 and mutants of the mGlu2 residues involved in JNJ-40411813 binding. n–p, LY354740-induced IP accumulation assay of WT mGlu2 and mutants of the mGlu2 residues involved in dimerization. r–t, Glutamate- and VU0364770-induced IP accumulation assay of WT mGlu4 and mutants of the mGlu4 residues involved in dimerization. Data are mean ± s.e.m. from at least five independent experiments performed in technical triplicate (IP accumulation) or duplicate (NanoBiT). Extended Data Tables 2, 4 provide detailed numbers of independent experiments (n) and statistical evaluation of the IP accumulation data. Extended Data Table 3 provides detailed numbers of independent experiments (n) and statistical evaluation of the NanoBiT data. u, Crosslinking of the mGlu2 mutants C121A/P7786.55C and C121A/V7967.34C as well as C121A/Y7816.58C and C121A/W5671.39C with (+) or without (−) pre-incubation with the agonist LY354740. The WT receptor and the C121A mutant were evaluated in parallel as controls. Three independent experiments were performed with similar results. Results from a representative experiment are shown. For gel source data, see Supplementary Fig. 1. v, Basal activity of the WT mGlu2 and mutants of residues involved in asymmetric dimerization. The IP accumulation assays were performed in parallel with the measurement of the IP production using the cells transfected only with the chimeric Gα protein Gαqi9 as a control. The basal activity was calculated by subtracting the IP production measured in the control for the WT receptor and all the mutants and is shown as per cent of the activity of the C121A mutant. The basal activity was substantially reduced by adding the orthosteric antagonist LY341495 (ref. 48). Data are mean ± s.e.m. (bars) with individual data points shown (dots). The numbers of independent experiments (n) performed in technical triplicate are shown in the parentheses. ***P < 0.0001 by one-way analysis of variance (ANOVA) followed by Dunnett’s post-test, compared with the basal activity of C121A (WT, P = 0.9994; WT–LY341495, F747C–H815C, R750C–H815C, I771C–V804C, L774C–L800C, P778C–V796C and Y781C–W567C, P < 0.0001). #The mutation C121A was introduced in both subunits for all the mutants. Protein expression levels of the mGlu2 mutants at the cell surface were determined in parallel by flow cytometry with an anti-Flag antibody (Sigma, 1:100 diluted in TBS + 4% BSA) and reported as per cent compared to the C121A mutant (% of C121A) from at least three independent measurements performed in technical duplicate: WT, 103 ± 1; F747C–H815C, 115 ± 5; R750C–H815C, 106 ± 5; I771C–V804C, 90 ± 3; L774C–L800C, 96 ± 10; P778C–V796C, 102 ± 7; Y781C–W567C, 88 ± 1.
Extended Data Fig. 6 Sequence alignment of the TMDs in the human mGlus and GABAB.
Colours represent the similarity of residues: red background, identical; red text, strongly similar. The mGlu2 residues that are involved in Gi binding are indicated by red arrows. The mGlu2 residues that mediate dimerization in the mGlu2–Gi1 structure are indicated by green arrows, and labelled with ‘G’ (in mGlu2G) and/or ‘f’ (in mGlu2free). The alignment was generated using UniProt (http://www.uniprot.org/align/) and the graphic was prepared on the ESPript 3.0 server (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi).
Supplementary information
Supplementary Figure
Uncropped blots for Extended Data Figures 1 and 5.
Rights and permissions
About this article
Cite this article
Lin, S., Han, S., Cai, X. et al. Structures of Gi-bound metabotropic glutamate receptors mGlu2 and mGlu4. Nature 594, 583–588 (2021). https://doi.org/10.1038/s41586-021-03495-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03495-2
This article is cited by
-
G protein-coupled receptors (GPCRs): advances in structures, mechanisms, and drug discovery
Signal Transduction and Targeted Therapy (2024)
-
Constitutive activation mechanism of a class C GPCR
Nature Structural & Molecular Biology (2024)
-
Allosteric modulation and G-protein selectivity of the Ca2+-sensing receptor
Nature (2024)
-
A framework for Frizzled-G protein coupling and implications to the PCP signaling pathways
Cell Discovery (2024)
-
Structure, function and drug discovery of GPCR signaling
Molecular Biomedicine (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.