Abstract
Crohn’s disease is a chronic inflammatory intestinal disease that is frequently accompanied by aberrant healing and stricturing complications. Crosstalk between activated myeloid and stromal cells is critical in the pathogenicity of Crohn’s disease1,2, and increases in intravasating monocytes are correlated with a lack of response to anti-TNF treatment3. The risk alleles with the highest effect on Crohn’s disease are loss-of-function mutations in NOD24,5, which increase the risk of stricturing6. However, the mechanisms that underlie pathogenicity driven by NOD2 mutations and the pathways that might rescue a lack of response to anti-TNF treatment remain largely uncharacterized. Here we use direct ex vivo analyses of patients who carry risk alleles of NOD2 to show that loss of NOD2 leads to dysregulated homeostasis of activated fibroblasts and macrophages. CD14+ peripheral blood mononuclear cells from carriers of NOD2 risk alleles produce cells that express high levels of collagen, and elevation of conserved signatures is observed in nod2-deficient zebrafish models of intestinal injury. The enrichment of STAT3 regulation and gp130 ligands in activated fibroblasts and macrophages suggested that gp130 blockade might rescue the activated program in NOD2-deficient cells. We show that post-treatment induction of the STAT3 pathway is correlated with a lack of response to anti-TNF treatment in patients, and demonstrate in vivo in zebrafish the amelioration of the activated myeloid–stromal niche using the specific gp130 inhibitor bazedoxifene. Our results provide insights into NOD2-driven fibrosis in Crohn’s disease, and suggest that gp130 blockade may benefit some patients with Crohn’s disease—potentially as a complement to anti-TNF therapy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data and supporting findings of this study are available within the Article and its Supplementary Information. All scRNA-seq data for human ileal samples have been deposited in the Gene Expression Omnibus (GEO) repository with accession code GSE134809. All 10X Chromium zebrafish scRNA-seq data have been deposited in the GEO repository with accession code GSE150498. The previously published34 dataset can also be found in the GEO repository with accession code GSE16879. The ZFIN genomic databases can be found at https://zfin.atlassian.net/wiki/spaces/general/pages/1891412257/Genomic+Resources+for+Zebrafish. Source data are provided with this paper.
Code availability
Scripts to reproduce zebrafish scRNA-seq clustering and differential expression analyses, as well as any other scripts used to generate computational figures, are available on GitHub at https://github.com/Cho-lab-Sinai/Scripts_Nayar_et_al.
References
Kugathasan, S. et al. Prediction of complicated disease course for children newly diagnosed with Crohn’s disease: a multicentre inception cohort study. Lancet 389, 1710–1718 (2017).
West, N. R. et al. Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nat. Med. 23, 579–589 (2017).
Martin, J. C. et al. Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell 178, 1493–1508.e20 (2019).
Ogura, Y. et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 411, 603–606 (2001).
Hugot, J. P. et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 411, 599–603 (2001).
Lesage, S. et al. CARD15/NOD2 mutational analysis and genotype-phenotype correlation in 612 patients with inflammatory bowel disease. Am. J. Hum. Genet. 70, 845–857 (2002).
Zhou, X. et al. Circuit design features of a stable two-cell system. Cell 172, 744–757.e17 (2018).
Verstockt, B. & Cleynen, I. Genetic influences on the development of fibrosis in Crohn’s disease. Front. Med. 3, 24 (2016).
Vermeire, S. et al. NOD2/CARD15 does not influence response to infliximab in Crohn’s disease. Gastroenterology 123, 106–111 (2002).
Mascheretti, S. et al. Pharmacogenetic investigation of the TNF/TNF-receptor system in patients with chronic active Crohn’s disease treated with infliximab. Pharmacogenomics J. 2, 127–136 (2002).
Brant, S. R. et al. Defining complex contributions of NOD2/CARD15 gene mutations, age at onset, and tobacco use on Crohn’s disease phenotypes. Inflamm. Bowel Dis. 9, 281–289 (2003).
Bain, C. C. et al. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat. Immunol. 15, 929–937 (2014).
Schafer, S. et al. IL-11 is a crucial determinant of cardiovascular fibrosis. Nature 552, 110–115 (2017).
Lim, W.-W. et al. Transgenic interleukin 11 expression causes cross-tissue fibro-inflammation and an inflammatory bowel phenotype in mice. PLoS ONE 15, e0227505 (2020).
Kim, H.-S. et al. Identification of novel Wilms’ tumor suppressor gene target genes implicated in kidney development. J. Biol. Chem. 282, 16278–16287 (2007).
Tamplin, O. J. et al. Hematopoietic stem cell arrival triggers dynamic remodeling of the perivascular niche. Cell 160, 241–252 (2015).
Chuang, L.-S. et al. Zebrafish modeling of intestinal injury, bacterial exposures and medications defines epithelial in vivo responses relevant to human inflammatory bowel disease. Dis. Model. Mech. 12, dmm037432 (2019).
Kim, Y.-G. et al. The Nod2 sensor promotes intestinal pathogen eradication via the chemokine CCL2-dependent recruitment of inflammatory monocytes. Immunity 34, 769–780 (2011).
Reilkoff, R. A., Bucala, R. & Herzog, E. L. Fibrocytes: emerging effector cells in chronic inflammation. Nat. Rev. Immunol. 11, 427–435 (2011).
Sun, H. et al. Netrin-1 regulates fibrocyte accumulation in the decellularized fibrotic scleroderma lung microenvironment and in bleomycin induced pulmonary fibrosis. Arthritis Rheumatol. 68, 1251–1261 (2016).
García-de-Alba, C. et al. Expression of matrix metalloproteases by fibrocytes: possible role in migration and homing. Am. J. Respir. Crit. Care Med. 182, 1144–1152 (2010).
Girardin, S. E. et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem. 278, 8869–8872 (2003).
Orekhov, A. N. et al. Monocyte differentiation and macrophage polarization. Vessel Plus 3, 10 (2019).
Ma, G. et al. Nod2–Rip2 signaling contributes to intestinal injury induced by muramyl dipeptide via oligopeptide transporter in rats. Dig. Dis. Sci. 60, 3264–3270 (2015).
Strober, W., Kitani, A., Fuss, I., Asano, N. & Watanabe, T. The molecular basis of NOD2 susceptibility mutations in Crohn’s disease. Mucosal Immunol. 1, S5–S9 (2008).
Liu, Y. et al. Circulating fibrocytes are involved in inflammation and leukocyte trafficking in neonates with necrotizing enterocolitis. Medicine (Baltimore) 96, e7400 (2017).
Ishida, M. et al. Muramyl dipeptide enhances lipopolysaccharide-induced osteoclast formation and bone resorption through increased RANKL expression in stromal cells. J. Immunol. Res. 2015, 132765 (2015).
Kettleborough, R. N. W. et al. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature 496, 494–497 (2013).
Chassaing, B., Aitken, J. D., Malleshappa, M. & Vijay-Kumar, M. Dextran sulfate sodium (DSS)-induced colitis in mice. Curr. Protoc. Immunol. 104, Unit 15.25 (2014).
Waldner, M. J. & Neurath, M. F. Master regulator of intestinal disease: IL-6 in chronic inflammation and cancer development. Semin. Immunol. 26, 75–79 (2014).
Thilakasiri, P. et al. Repurposing the selective estrogen receptor modulator bazedoxifene to suppress gastrointestinal cancer growth. EMBO Mol. Med. 11, e9539 (2019).
Wei, J. et al. Bazedoxifene as a novel GP130 inhibitor for colon cancer therapy. J. Exp. Clin. Cancer Res. CR 38, 63 (2019).
Wu, X., Cao, Y., Xiao, H., Li, C. & Lin, J. Bazedoxifene as a novel GP130 inhibitor for pancreatic cancer therapy. Mol. Cancer Ther. 15, 2609–2619 (2016).
Arijs, I. et al. Mucosal gene expression of antimicrobial peptides in inflammatory bowel disease before and after first infliximab treatment. PLoS ONE 4, e7984 (2009).
Yeo, S.-Y. et al. A positive feedback loop bi-stably activates fibroblasts. Nat. Commun. 9, 3016 (2018).
Chen, K. L. A., Zhao, Y. C., Hieronymi, K., Smith, B. P. & Madak-Erdogan, Z. Bazedoxifene and conjugated estrogen combination maintains metabolic homeostasis and benefits liver health. PLoS ONE 12, e0189911 (2017).
Sontake, V. et al. Wilms’ tumor 1 drives fibroproliferation and myofibroblast transformation in severe fibrotic lung disease. JCI Insight 3, e121252 (2018).
Kinchen, J. et al. Structural remodeling of the human colonic mesenchyme in inflammatory bowel disease. Cell 175, 372–386.e17 (2018).
Buechler, M. B. et al. A stromal niche defined by expression of the transcription factor WT1 mediates programming and homeostasis of cavity-resident macrophages. Immunity 51, 119–130.e5 (2019).
Frei, R. et al. Early initiation of anti-TNF is associated with favourable long-term outcome in Crohn’s disease: 10-year-follow-up data from the Swiss IBD cohort study. J. Crohn’s Colitis 13, 1292–1301 (2019).
Danese, S. et al. Randomised trial and open-label extension study of an anti-interleukin-6 antibody in Crohn’s disease (ANDANTE I and II). Gut 68, 40–48 (2019).
Sands, B. E. et al. Randomized, controlled trial of recombinant human interleukin-11 in patients with active Crohn’s disease. Aliment. Pharmacol. Ther. 16, 399–406 (2002).
Viennois, E., Chen, F., Laroui, H., Baker, M. T. & Merlin, D. Dextran sodium sulfate inhibits the activities of both polymerase and reverse transcriptase: lithium chloride purification, a rapid and efficient technique to purify RNA. BMC Res. Notes 6, 360 (2013).
Zhang, X. et al. Luminally polarized mural and vascular remodeling in ileal strictures of Crohn’s disease. Hum. Pathol. 79, 42–49 (2018).
Dehairs, J., Talebi, A., Cherifi, Y. & Swinnen, J. V. CRISP-ID: decoding CRISPR mediated indels by Sanger sequencing. Sci. Rep. 6, 28973 (2016).
Haberman, Y. et al. Pediatric Crohn’s disease patients exhibit specific ileal transcriptome and microbiome signature. J. Clin. Invest. 124, 3617–3633 (2014).
Gettler, L. et al. Prioritizing Crohn’s disease genes by integrating association signals with gene expression implicates monocyte subsets. Genes Immun. 20, 577–588 (2019).
Love, M. I. et al. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Acknowledgements
We thank S. Grossman of the Dr Sanford J. Grossman Center for Integrative Studies in IBD, which is supported by the generosity of The Sanford J. Grossman Charitable Trust; the ISMMS Department of Pathology for providing human tissue samples; D. Derose, K. Svoboda, C. Chasteau and U. Korie for sample collection and coordination; A. Saha and H. Wen for recall of healthy volunteers with NOD2 mutations via the ISMMS BioMe Biobank; F. Avila and M. Garcia-Barros from the Biorepository and Pathology core for assistance with staining human ileal strictures; N. Hsu and N. Villaverde (members of the laboratory of J.H.C.) for discussions; and F. Marlow and B. Rosenberg for discussions relating to zebrafish and single-cell experiments. This work was also supported by National Institutes of Health (NIH) grants R01 DK106593, R01 DK123758-01 and U01 DK062422 to J.H.C.
Author information
Authors and Affiliations
Contributions
J.H.C. and S.N. conceived the project. S.N., J.K.M. and J.H.C. designed the experiments, and S.N. and J.K.M performed them. S.N. and J.H.C. wrote the manuscript, with discussions and edits from E.K., L.-s.C., J.C. and J.K.M. L.-s.C. and M.M. provided intellectual input throughout the study’s progression. M.G. and K.G. provided substantial assistance with the generation of computational analyses. S.K. and K.G. assisted in the handling of genetic and clinical data from the RISK dataset. L.A.W. and E.K. provided assistance with scRNA-seq experiments and analysis strategies. H.M.K. provided human ileal stricture slides as well as expertise on histological analyses.
Corresponding author
Ethics declarations
Competing interests
S.N. and J.H.C., through the Icahn School of Medicine at Mount Sinai, have filed a provisional US patent application (no. 63/130,035) on repurposing BZA for clinical use in a subset of patients with Crohn’s disease. All other authors declare no competing interests.
Additional information
Peer review information Nature thanks Alison Simmons and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 scRNA-seq exploration of ileum of patients with Crohn’s disease and validation by protein staining.
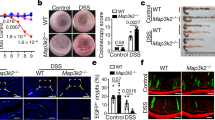
a, Genes (columns) across myeloid and stromal cells (rows) from scRNA-seq of uninflamed ileum of patients with Crohn’s disease (n = 11). Expression defined as log2(gene expression/average) across all shown clusters. b, Relative expression of NOD2, RIPK2 and XIAP (columns) across myeloid and stromal cells (rows) from scRNA-seq of PBMCs and inflamed ileum of patients with Crohn’s disease (n = 11). c, Gene ontology analysis from upregulated genes in activated fibroblasts from inflamed ileum. The number of genes per biological process is shown in parentheses. Processes ranked from top to bottom in decreasing order of −log(P value). d, Percentage of COL1A1+, PDGFRA+, CD14+ and CD14+PDGFRA+ double-positive cells of total activated fibroblasts (1,367 cells total) in inflamed ileum. e, Number of cells from direct ex vivo sorting of CD14+PDGFRA+ cells of uninflamed and inflamed ileum of patients with Crohn’s disease. n = 3 biological replicates. Data are mean ± s.e.m. f, Full-thickness sections of inflamed strictures stained with MPEG1 and MFAP4. n = 3 biological replicates per stain. g, Full-thickness sections of inflamed strictures showing expression of WT1, PDGFRA and CD14. n = 2 patients per stain (patient 7 and 8 denoted from a previous publication3) (left). In inflamed strictures from samples from patients with Crohn’s disease, WT1, PDGFRA and CD14 expression is seen in muscularis mucosae and lymphoid aggregates; and around blood vessels (right). n = 2 patients per stain; 5 images per patient. Scale bar, approximately 50 μm. h, RT–PCR of gene expression at 24 h after 1× DSS-treatment of zebrafish nod2+/+ and nod2sa21011/sa21011 larvae, comparing intestinal and carcass expression. n = 3 biological replicates; 3 clutches, 10–15 larvae per genotype per clutch. Data are mean ± s.e.m. *P < 0.05, **P < 0.01; two-tailed paired Student’s t-test.
Extended Data Fig. 2 scRNA-seq data from intestines of DSS-treated zebrafish larvae.
a, scRNA-seq of intestines of untreated and DSS-treated zebrafish larvae from nod2+/+ and nod2mss13/mss13 backgrounds. UMAP shows joint clustering of 30,069 cells, revealing 32 unique clusters (left). Top 5 markers for each cluster (rows) (right). b, UMAP showing human–zebrafish clusters from species integration via scRNA-seq (top); top 3 markers for each cluster (rows) (bottom). Expression is UMI counts per gene per cell. c, Homologous expression between zebrafish and human clusters (expression is log-normalized).
Extended Data Fig. 3 NOD2 deficiency enhances differentiation of spindle cells from CD14+ monocytes.
a, Morphological quantification of PBMC differentiation from NOD2WT/WT or NOD2MT/MT cells, with 2.5 μg ml−1 Pam3Cys, 0.5 μg ml−1 lipid A and 1 μg ml−1 MDP. Classification of categories in Methods. n = 6 biological replicates for NOD2WT/WT, n = 3 biological replicates for NOD2MT/MT for each treatment condition. Data are mean ± s.e.m. *P < 0.05, **P < 0.01; two-sided Wilcoxon signed-rank t-test. b, Representative morphological images of spindle, intermediate and round cells from NOD2WT/WT, NOD2WT/MT and NOD2MT/MT individuals, unstimulated or treated with 1 μg ml−1 MDP, 0.5 μg ml−1 lipid A or 2.5 μg ml−1 Pam3Cys. n = 5 images per well condition per patient. Scale bar, 100 μm. c, Representative images of myeloid (MPEG1) and stromal (MFAP4, COL5A1 and COL1A1) expression by immunofluorescence (left). Quantification of staining (right). Data are mean ± s.e.m. of corrected cellular fluorescence. ***P < 0.001; two-way ANOVA test with Sidak correction. n = 4 images per fluorescent marker; individual values, individual cells per image. d, Schematic of CD14+CD16− PBMCs isolated from healthy NOD2-mutant carriers and non-carriers, and differentiation assay (left). RT–PCR of gene expression from 1 μg ml−1 MDP-treated PBMCs after 2 weeks of differentiation from NOD2WT/WT or NOD2MT/MT individuals. n = 3 biological replicates (right). Data are mean MDP relative expression (left y axis) and individual 2−ΔCt expression values (right y axis) with error bars representing individual ΔCt values ± s.e.m.. e, RT–PCR of gene expression of unstimulated CD14+CD16+ PBMCs after two weeks of differentiation from NOD2WT/WT or NOD2MT/MT individuals. n = 3 biological replicates. Data are mean untreated relative expression (left y axis) and individual 2−ΔCt expression values (right y axis) with error bars representing individual ΔCt values ± s.e.m. f, Secreted protein (pg ml−1) by Luminex from unstimulated PBMCs or PBMCs stimulated with 0.5 μg ml−1 lipid A, 2.5 μg ml−1 Pam3Cys or 1 μg ml−1 MDP. n = 2 NOD2WT/WT, n = 3 NOD2MT/MT. Data are mean ± s.e.m. *P < 0.05; two-way ANOVA. g, Schematic of nod2sa21011 mutant from Zebrafish Sanger Mutation Project and nod2mss13/mss/13 CRISPR-knockout zebrafish line (left); timeline of 1× and 2× MDP stimulation of zebrafish larvae (right).
Extended Data Fig. 4 Staining of key fibrotic protein deposition in zebrafish larvae and human ileal stricture resections.
a, Nod2 expression as assessed by western blotting to show loss of protein levels in nod2sa21011 and nod2mss13/mss13 CRISPR-mutant zebrafish larvae. Larvae were untreated and protein was collected at 6 dpf. n = 1 biological replicate per nod2 mutation for protein-knockout validation (1 clutch per mutation line with 20 larvae for each genotype). For gel source data, see Supplementary Fig. 1. b, UMAP of myeloid and stromal clusters from joint clustering of scRNA-seq data for zebrafish larval cells, grouped by nod2+/+ and nod2mss13/mss13 genotypes. c, NOD2 mutation information of patients in the ileal Crohn’s disease scRNA-seq cohort, used for differential expression analysis between activated and nonactivated fibroblast and macrophage clusters. d, Transcription factors from ingenuity pathway analysis upstream of genes enriched in activated fibroblasts and inflammatory macrophages from individuals with NOD2 risk alleles versus individuals without these alleles. P values were determined by ingenuity pathway analysis for transcription-factor regulation of genes in differential expression analysis. e, Top 5 genes (rows) in each cluster (columns). Expression is in UMI counts. f, Feature plots to show localized expression of key transcripts in nod2WT/WT larvae from scRNA-seq myeloid and stromal populations (log-normalized expression).
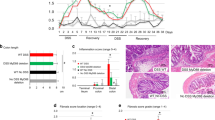
Extended Data Fig. 5 gp130 blockade by BZA ameliorates fibrotic activation and may provide a supplementary approach to anti-TNF therapy.
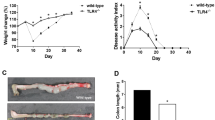
a, log2-transformed microarray expression from a previous publication34, of gp130-associated genes in patients treated with infliximab. Blue bars, patients who responded to infliximab (n = 18); red bars, patients who did not respond to infliximab (n = 16). Data are mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; two-tailed paired Student’s t-test. b, Morphological quantification of NOD2WT/WT or NOD2MT/MT-differentiated PBMCs that were unstimulated or stimulated with 0.5 μg ml−1 MDP or 0.5 μg ml−1 MDP + 1 μM BZA. n = 6 biological replicates for NOD2WT/WT (n = 4 for MDP + BZA costimulation) and n = 3 biological replicates for NOD2MT/MT per each treatment condition (top); unstimulated, 0.5 μg ml−1 MDP, 0.5 μg ml−1 MDP + 1 μM BZA or MDP + 0.05 μg ml−1 monoclonal anti-gp130 antibody (bottom). n = 2 biological replicates for each genotype. Data are mean ± s.e.m. *P < 0.05, **P < 0.01; two-sided Wilcoxon signed-rank test. c, Secreted protein (pg ml−1) by Luminex from unstimulated PBMCs or PBMCs stimulated with 0.5 μg ml−1 MDP, 0.5 μg ml−1 MDP + 1 μM BZA or 0.5 μg ml−1 MDP + 0.05 μg ml−1 anti-gp130. n = 2 individuals per genotype. data are mean values. d, BZA target-gene expression in stromal and myeloid cells of scRNA-seq data from DSS-treated larvae (log-normalized expression). e, Percentage of stromal and myeloid populations in total intestinal cells sequenced in DSS-treated versus DSS + BZA-treated larvae. Red boxes, nod2mss13/mss13 differences. f, g, Violin plots for upregulated genes upon DSS treatment, or for genes downregulated by DSS + BZA cotreatment in nod2+/+ (f) and nod2mss13/mss13 (g) larvae (log-normalized expression). h, Haematoxylin and eosin staining of DSS-treated and DSS + BZA-treated nod2+/+ and nod2mss13/mss13 zebrafish larvae. Intestinal bulb hypertrophy measured as indicated by scale bars, and quantified after 1× (left) and 2× (right) treatments. n = 5–10 biological replicates (individual larvae) per condition. Data are mean ± s.e.m. For nod2+/+, 5 untreated (1×), 3 1×DSS, 4 untreated (2×), 10 2×DSS; for nod2mss13, n = 6 untreated (1×, 8 1×DSS, 5 untreated (2×), 5 2×DSS. *P < 0.05, **P < 0.01; one way-ANOVA with Tukey’s multiple comparison. i, Intestine length of 2×DSS-treated or 2×DSS + BZA-cotreated nod2+/+, nod2+/− and nod2−/− larvae. n = 3 clutches; 10–15 larvae per group. Data are mean ± s.e.m. *P < 0.05, **P < 0.01; two-way ANOVA test with Tukey’s multiple comparison. j, RT–PCR of gene expression at 24 h after 1× or 2×DSS + BZA cotreatment of nod2+/+, nod2+/− or nod2−/− zebrafish larvae. n = 4 biological replicates for 1× treatment; 4 clutches, 10–15 larvae per genotype per clutch. n = 4 biological replicates for 2× treatment; 4 clutches, 10–15 larvae per genotype per clutch. Data are mean ± s.e.m.
Extended Data Fig. 6 Pathogenesis of Crohn’s disease and treatment pathways.
Model to show proposed mechanism of pathophysiology of Crohn’s disease under specific genetic and cellular constraints. Individuals who carry a NOD2 risk allele develop aberrant fibroblast–macrophage homeostasis and differentiation over time. At preclinical stages of disease development, patients with Crohn’s disease increase production of antibodies (ASCA and CBir) to contain infection caused by increased bacterial load and elevate inflammatory mediators (cytokines and chemokines such as IL11, CXCL13, IL6, CCL2 and OSM). Patients who carry NOD2 risk alleles will upregulate a pathogenic activated fibroblast and macrophage signature with disease development. This results in downstream activation of STAT3, among other chronic inflammatory and fibrotic consequences that we have shown in this Article. Some patients administered anti-TNF therapy will not respond, and often develop complications such as stricturing disease. Furthermore, over time, initial primary responders may become secondary nonresponders. Here we show that patients who are treatment-refractory have increased gp130 and activated fibroblast signatures; these would be patients for whom supplementing anti-TNF treatment with the gp130 inhibitor BZA could be beneficial. This model summarizes a two-step candidate selection approach: first by patients with Crohn’s disease who carrying NOD2 risk alleles with elevation of key expression signatures, and then by a lack of response to anti-TNF) to inform personalized therapeutic decision-making for Crohn’s disease.
Supplementary information
Supplementary Figure 1
Source gels for Extended Data Figure 4a. Top gel showing protein expression of Nod2 and tubulin control from nod2sa21011 mutant line. Bottom gel showing protein expression of Nod2 and tubulin control from CRISPR-mediated knockout nod2mss13 line. Note that in the bottom gel the homozygote fish lysate is loaded next to the WT lysate, followed by the heterozygote fish lysate. Protein markers are directly overlaid from the BioRad Precision Plus Protein ladder. Red boxes indicate portions of the gels displayed in Extended Data Fig. 4a.
Supplementary Table 1
UMI counts of all cells and genes in single-cell ileal dataset. Table to illustrate unique molecular identifier counts per gene per cell per sample (as shown by nCount_RNA). Object indicates sample ID (uninflamed or inflamed ileal tissue from an individual). Supplementary metadata for each tissue sample can be found in Martin et al., 2019. Group indicates tissue inflammation status. Cluster indicates barcode and cell designation to a specific single-cell cluster.
Supplementary Table 2
UMI counts of CD14+PDGFRA+ cells in single-cell ileal dataset. Table to illustrate unique molecular identifier counts CD14+PDGFRA+ double-expressing cells per sample (as shown by nCount_RNA). Object indicates sample ID (uninflamed or inflamed ileal tissue from an individual). Group indicates tissue inflammation status. Cluster indicates barcode and cell designation to a specific single-cell cluster; here, cluster 20 is designated as “activated fibroblasts”, which is the cell type that is predominantly expressing CD14+ and PDGFRA+ in the same cell. UMI distribution does not fall out of the range of all UMIs across all genes and cell types, indicating that these CD14+PDGFRA+ cells are single cells co-expressing both genes, as opposed to doublets.
Supplementary Table 3
Cell-type specific marker expression for all cells in zebrafish single-cell RNA sequencing dataset. Table to illustrate top differentially expressed genes in each cluster for zebrafish single-cell RNA sequencing (joint clustering model; sheet 1) and zebrafish + human scRNAseq integrated model (joint clustering; sheet 2). Analyses were performed using the function “FindAllMarkers()” in Seurat. v3, s. pct.1 shows percent of all cells expressing the gene within that cluster and pct.2 shows percent of all cells expressing the gene in all other clusters being compared in the comparison.
Supplementary Table 4
Differentially expressed genes between activated and residential fibroblast and macrophage populations in inflamed ileal CD scRNA seq data. Table to illustrate top differentially expressed genes between a) activated fibroblasts vs. residential fibroblasts and b) inflammatory macrophages vs. residential macrophages in scRNA sequencing of 11 inflamed CD ileum cells. “FindAllMarkers()” in Seurat. v3, which uses a two-sided Wilcoxon test to perform differential gene expression analysis. pct.1 shows percent of all cells expressing the gene within activated/inflamed cluster and pct.2 shows percent of all cells expressing the gene in residential clusters being compared in the analysis.
Supplementary Table 5
PCA generation of “activated fibroblasts” and “inflammatory macrophages” correlated with NOD2 risk allele carriers in RISK cohort. Table illustrating top principal components derived from top 200 differentially expressed genes in activated fibroblasts and inflammatory macrophages across each patient in pediatric inception RISK cohort. Genes are derived from differential expression analysis between activated and non-activated fibroblasts and macrophages from single-cell RNA seq ileal dataset (ref. Supplementary Table 4).
Supplementary Table 6
Regression analyses output for NOD2 allele status and stricture development in RISK cohort. Series of regression tests to test relationships between NOD2 risk alleles and stricture phenotype in RISK cohort. Using all 8 associated SNPs, rs2066847 frameshift NOD2 mutation is significant in predicting stricture status; using total NOD2 minor allele count, the sum of NOD2 risk alleles is significant in predicting stricture status; 0 vs. 1, 0 vs.2 NOD2 alleles to predict stricture status show increasing levels of significance with increasing number of NOD2 risk alleles.
Supplementary Table 7
Cell type-specific marker expression for stromal and myeloid cells only in zebrafish single-cell RNA sequencing dataset. Table to illustrate top differentially expressed genes in each cluster for stromal and myeloid cells only in zebrafish single-cell RNA sequencing (joint clustering model). Analyses were performed using the function “FindAllMarkers()” in Seurat. v3, using a two-sided negative binomial test to perform differential gene expression analysis. pct.1 shows percent of all cells expressing the gene within that cluster and pct.2 shows percent of all cells expressing the gene in all other clusters being compared in the comparison.
Supplementary Table 8
Differential expression analysis between activated fibroblasts and inflammatory macrophages in NOD2 risk allele carriers from inflamed ileal single-cell RNA seq dataset. Table to show differential expression analysis performed in Seurat v3 between tab 1) activated and non-activated fibroblasts and tab 2) inflammatory and non-inflammatory macrophages between NOD2 risk allele carriers (N=3) and NOD2 wildtype carriers (N=4). pct.1 indicates percent of cells of NOD2 risk allele carriers expressing specific gene, and pct.2 indicates percent of cells of NOD2 wildtype patients expressing specific gene. two-sided Wilcoxon test parameter in Seurat 3 used.
Supplementary Table 9
Ingenuity Pathway Analysis output of transcriptional regulators upstream of differentially expressed genes in activated fibroblasts and inflammatory macrophages of NOD2 risk allele carriers. Supplementary Tables 8 (gene, logFC, adj-p-value) were used as input data for performing Ingenuity Pathway Analysis. Results for upstream transcriptional regulators are shown, along with their target molecules that are a subset of the input differential expression geneset. Transcription factors highlighted are those we here implicate play key roles in orchestrating chronic inflammation and fibrosis in Crohn’s disease.
Supplementary Table 10
Patient characteristics for NOD2 healthy volunteer recall. Clinical and demographic information for each healthy volunteer who participated in blood draw for PBMC isolation. NOD2 genotype illustrates the mutation status of each patient, and DMSEA indicates all patients were of European Ancestry. An age cutoff of 65 years old was used.
Rights and permissions
About this article
Cite this article
Nayar, S., Morrison, J.K., Giri, M. et al. A myeloid–stromal niche and gp130 rescue in NOD2-driven Crohn’s disease. Nature 593, 275–281 (2021). https://doi.org/10.1038/s41586-021-03484-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03484-5
This article is cited by
-
Combination of bazedoxifene with chemotherapy and SMAC-mimetics for the treatment of colorectal cancer
Cell Death & Disease (2024)
-
Intestinal stroma guides monocyte differentiation to macrophages through GM-CSF
Nature Communications (2024)
-
The role of admixture in the rare variant contribution to inflammatory bowel disease
Genome Medicine (2023)
-
Macrophages in intestinal homeostasis and inflammatory bowel disease
Nature Reviews Gastroenterology & Hepatology (2023)
-
A CCL2+DPP4+ subset of mesenchymal stem cells expedites aberrant formation of creeping fat in humans
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.