Abstract
Symmetric cell division requires the even partitioning of genetic information and cytoplasmic contents between daughter cells. Whereas the mechanisms coordinating the segregation of the genome are well known, the processes that ensure organelle segregation between daughter cells remain less well understood1. Here we identify multiple actin assemblies with distinct but complementary roles in mitochondrial organization and inheritance in mitosis. First, we find a dense meshwork of subcortical actin cables assembled throughout the mitotic cytoplasm. This network scaffolds the endoplasmic reticulum and organizes three-dimensional mitochondrial positioning to ensure the equal segregation of mitochondrial mass at cytokinesis. Second, we identify a dynamic wave of actin filaments reversibly assembling on the surface of mitochondria during mitosis. Mitochondria sampled by this wave are enveloped within actin clouds that can spontaneously break symmetry to form elongated comet tails. Mitochondrial comet tails promote randomly directed bursts of movement that shuffle mitochondrial position within the mother cell to randomize inheritance of healthy and damaged mitochondria between daughter cells. Thus, parallel mechanisms mediated by the actin cytoskeleton ensure both equal and random inheritance of mitochondria in symmetrically dividing cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Source data are provided with this paper.
Code availability
MATLAB code used for analysing 3D mitochondrial distribution, 2D nucleoid distribution, ER void size and distribution, mitochondrial motility and damaged-mitochondria circular spread is available at https://github.com/andmoo91/CircularAnalyses_MatLab.
References
Carlton, J. G., Jones, H. & Eggert, U. S. Membrane and organelle dynamics during cell division. Nat. Rev. Mol. Cell Biol. 21, 151–166 (2020).
Taguchi, N., Ishihara, N., Jofuku, A., Oka, T. & Mihara, K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J. Biol. Chem. 282, 11521–11529 (2007).
Kashatus, D. F. et al. RALA and RALBP1 regulate mitochondrial fission at mitosis. Nat. Cell Biol. 13, 1108–1115 (2011).
Chung, J. Y., Steen, J. A. & Schwarz, T. L. Phosphorylation-induced motor shedding is required at mitosis for proper distribution and passive inheritance of mitochondria. Cell Rep. 16, 2142–2155 (2016).
Altmann, K., Frank, M., Neumann, D., Jakobs, S. & Westermann, B. The class V myosin motor protein, Myo2, plays a major role in mitochondrial motility in Saccharomyces cerevisiae. J. Cell Biol. 181, 119–130 (2008).
Simon, V. R., Swayne, T. C. & Pon, L. A. Actin-dependent mitochondrial motility in mitotic yeast and cell-free systems: identification of a motor activity on the mitochondrial surface. J. Cell Biol. 130, 345–354 (1995).
Rohn, J. L. et al. Myo19 ensures symmetric partitioning of mitochondria and coupling of mitochondrial segregation to cell division. Curr. Biol. 24, 2598–2605 (2014).
Field, C. M. & Lénárt, P. Bulk cytoplasmic actin and its functions in meiosis and mitosis. Curr. Biol. 21, R825–R830 (2011).
Kita, A. M. et al. Spindle-F-actin interactions in mitotic spindles in an intact vertebrate epithelium. Mol. Biol. Cell 30, 1645–1654 (2019).
Salvador-Gallego, R., Hoyer, M. J. & Voeltz, G. K. SnapShot: functions of endoplasmic reticulum membrane contact sites. Cell 171, 1224–1224 (2017).
Lu, L., Ladinsky, M. S. & Kirchhausen, T. Cisternal organization of the endoplasmic reticulum during mitosis. Mol. Biol. Cell 20, 3471–3480 (2009).
Mitsushima, M. et al. Revolving movement of a dynamic cluster of actin filaments during mitosis. J. Cell Biol. 191, 453–462 (2010).
Moore, A. S., Wong, Y. C., Simpson, C. L. & Holzbaur, E. L. Dynamic actin cycling through mitochondrial subpopulations locally regulates the fission–fusion balance within mitochondrial networks. Nat. Commun. 7, 12886 (2016).
Taunton, J. et al. Actin-dependent propulsion of endosomes and lysosomes by recruitment of N-WASP. J. Cell Biol. 148, 519–530 (2000).
Kast, D. J., Zajac, A. L., Holzbaur, E. L., Ostap, E. M. & Dominguez, R. WHAMM directs the Arp2/3 complex to the ER for autophagosome biogenesis through an actin comet tail mechanism. Curr. Biol. 25, 1791–1797 (2015).
Tilney, L. G. & Portnoy, D. A. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J. Cell Biol. 109, 1597–1608 (1989).
Heinzen, R. A., Hayes, S. F., Peacock, M. G. & Hackstadt, T. Directional actin polymerization associated with spotted fever group Rickettsia infection of Vero cells. Infect. Immun. 61, 1926–1935 (1993).
Bernardini, M. L., Mounier, J., d’Hauteville, H., Coquis-Rondon, M. & Sansonetti, P. J. Identification of icsA, a plasmid locus of Shigella flexneri that governs bacterial intra- and intercellular spread through interaction with F-actin. Proc. Natl Acad. Sci. USA 86, 3867–3871 (1989).
Haglund, C. M., Choe, J. E., Skau, C. T., Kovar, D. R. & Welch, M. D. Rickettsia Sca2 is a bacterial formin-like mediator of actin-based motility. Nat. Cell Biol. 12, 1057–1063 (2010).
Theriot, J. A., Mitchison, T. J., Tilney, L. G. & Portnoy, D. A. The rate of actin-based motility of intracellular Listeria monocytogenes equals the rate of actin polymerization. Nature 357, 257–260 (1992).
Nishimura, Y. et al. The formin inhibitor, SMIFH2, inhibits members of the myosin superfamily. Preprint at https://doi.org/10.1101/2020.08.30.274613 (2020).
Manor, U., Bartholomew, S., Golani, G. & Christenson, E. A mitochondria-anchored isoform of the actin-nucleating spire protein regulates mitochondrial division. eLife 4, e08828 (2015).
Korobova, F., Ramabhadran, V. & Higgs, H. N. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science 339, 464–467 (2013).
Chhabra, E. S. & Higgs, H. N. INF2 Is a WASP homology 2 motif-containing formin that severs actin filaments and accelerates both polymerization and depolymerization. J. Biol. Chem. 281, 26754–26767 (2006).
Vincent, A. E. et al. Subcellular origin of mitochondrial DNA deletions in human skeletal muscle. Ann. Neurol. 84, 289–301 (2018).
Katajisto, P. et al. Stem cells. Asymmetric apportioning of aged mitochondria between daughter cells is required for stemness. Science 348, 340–343 (2015).
Aryaman, J., Hoitzing, H., Burgstaller, J. P., Johnston, I. G. & Jones, N. S. Mitochondrial heterogeneity, metabolic scaling and cell death. BioEssays 39, 1700001 (2017).
McFaline-Figueroa, J. R. et al. Mitochondrial quality control during inheritance is associated with lifespan and mother-daughter age asymmetry in budding yeast. Aging Cell 10, 885–895 (2011).
Rué, P. & Martinez Arias, A. Cell dynamics and gene expression control in tissue homeostasis and development. Mol. Syst. Biol. 11, 792 (2015).
Stewart, J. B. & Chinnery, P. F. Extreme heterogeneity of human mitochondrial DNA from organelles to populations. Nat. Rev. Genet. 22, 106–118 (2020).
Fernandopulle, M. S. et al. Transcription factor-mediated differentiation of human iPSCs into neurons. Curr. Protoc. Cell Biol. 79, e51 (2018).
Simpson, C. L., Kojima, S. & Getsios, S. RNA interference in keratinocytes and an organotypic model of human epidermis. Methods Mol. Biol. 585, 127–146 (2010).
Xu, K., Zhong, G. & Zhuang, X. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science 339, 452–456 (2013).
Chozinski, T. J. et al. Expansion microscopy with conventional antibodies and fluorescent proteins. Nat. Methods 13, 485–488 (2016).
Guo, Y. et al. visualizing intracellular organelle and cytoskeletal interactions at nanoscale resolution on millisecond timescales. Cell 175, 1430–1442 (2018).
Preibisch, S., Saalfeld, S., Schindelin, J. & Tomancak, P. Software for bead-based registration of selective plane illumination microscopy data. Nat. Methods 7, 418–419 (2010).
Chen, B.-C. et al. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science 346, 1257998 (2014).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Steger, C. An unbiased detector of curvilinear structures. IEEE Trans. Pattern Anal. Mach. Intell. 20, 113–125 (1998).
Püspöki, Z., Storath, M., Sage, D. & Unser, M. Transforms and operators for directional bioimage analysis: a survey. Adv. Anat. Embryol. Cell Biol. 219, 69–93 (2016).
Berens, P. CircStat: a MATLAB toolbox for circular statistics. J. Stat. Softw. 31, https://doi.org/10.18637/jss.v031.i10 (2009).
Upton, G. & Mardia, K. Statistics of directional data. J. R. Stat. Soc. A 136, 262–263 (1973).
Berg, S. et al. ilastik: interactive machine learning for (bio)image analysis. Nat. Methods 16, 1226–1232 (2019).
Wait, E., Winter, M. & Cohen, A. R. Hydra image processor: 5-D GPU image analysis library with MATLAB and Python wrappers. Bioinformatics 35, 5393–5395 (2019).
Tinevez, J.-Y. et al. TrackMate: an open and extensible platform for single-particle tracking. Methods 115, 80–90 (2017).
Tarantino, N. et al. TNF and IL-1 exhibit distinct ubiquitin requirements for inducing NEMO-IKK supramolecular structures. J. Cell Biol. 204, 231–245 (2014).
Monnier, N. et al. Bayesian approach to MSD-based analysis of particle motion in live cells. Biophys. J. 103, 616–626 (2012).
Weber, S. C., Thompson, M. A., Moerner, W. E., Spakowitz, A. J. & Theriot, J. A. Analytical tools to distinguish the effects of localization error, confinement, and medium elasticity on the velocity autocorrelation function. Biophys. J. 102, 2443–2450 (2012).
Quintero, O. A. et al. Human Myo19 is a novel myosin that associates with mitochondria. Curr. Biol. 19, 2008–2013 (2009).
Ortega, F. E., Koslover, E. F. & Theriot, J. A. Listeria monocytogenes cell-to-cell spread in epithelia is heterogeneous and dominated by rare pioneer bacteria. eLife 8, e40032 (2019).
Acknowledgements
We thank M. Tokito, A. Stout and the UPenn Cell and Developmental Biology microscopy core, and F. Tuluc and the CHOP Flow Cytometry core for technical contributions; S. Weber for MATLAB code; O. Quintero and D. Matus for cell lines; H. Choi and B. Rossetti for advice on analyses; and M. Ostap, F. Cross, M. Moore, C. Evans, S. Cason, Y. Wong and O. Harding for critical feedback. This work was funded by support from NIH R35 GM126950 and RM1 GM136511 to E.L.F.H., NIH F31 GM123644 and CHOP MitoRAG Pilot Grant to A.S.M., NIH R37 AI036929 and HHMI investigator support to J.A.T., and a HHMI Gilliam Fellowship to F.E.O. C.L.S. was supported by grants from NIAMS/NIH (K08 AR075846), the Dermatology Foundation, and the National Psoriasis Foundation. NHEKs were supplied by Penn SBDRC Core B supported by NIAMS/NIH (P30 AR069589). J.J.N. was supported by F30NS092227 and the UPenn NGG Hearst Fellowship. C.A.B. was supported by the German Research Foundation (DFG; BO 5434/1-1). J.L.-S. was supported by HHMI. The Advanced Imaging Center at the Janelia Research Campus is a jointly funded venture of the Gordon and Betty Moore Foundation and the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
A.S.M. and E.L.F.H. conceptualized the project and designed experiments. A.S.M performed imaging experiments, C.L.S. prepared and transduced isolated NHEK and organotypic NHEK cultures, S.M.C. performed western blots, J.M.H. assisted with lattice light-sheet experiments, P.G.-D. cultured and transfected primary hippocampal neurons, C.A.B. cultured and transfected iPS cells. A.S.M., J.J.N., E.C.W., S.M.C. and C.J.O. analysed data. E.C.W. designed custom image analysis suite to segment and visualize lattice light-sheet microscopy data. F.E.O. and J.A.T. designed and implemented simulations. A.S.M. designed the figures. T.-L.C., J.A.T. and J.L.-S. provided critical feedback. A.S.M. and E.L.F.H. wrote the paper and all authors contributed to revisions.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Benedikt Westermann and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
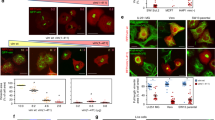
Extended Data Fig. 1 Mitochondrial networks are uniformly distributed throughout the cytoplasm surrounding the mitotic spindle.
a, z-coded xy and xz maximum-intensity projections of approximately 4-μm slabs through a live, metaphase HeLa cells expressing mito–dsRed2. Mitochondria are excluded from the spindle region and are uniformly distributed above, below, and around the spindle. b–d, z-coded xy and xz maximum-intensity projections of approximately 4 μm sections through the centre of A549 (b), HEK 293T (c) and iPS (d) cells expressing mito–dsRed2. e, z-coded xy (left) and xz (right) maximum-intensity projections of approximately 4-μm sections through the centre of a live, organotypic culture of NHEKs expressing mito–dsRed2. f, 4× expanded iSIM rendering of mitochondria (anti-HSP60) and microtubules (anti-α-tubulin) in a prometaphase HeLa cell. Mitochondria are entirely excluded from the spindle and show limited overlap with astral microtubules. Scale bars: 5 μm (a–d), 10 μm (e), 40 μm (f).
Extended Data Fig. 2 A 3D meshwork of actin cables associates with mitochondrial networks in the deep metaphase cytoplasm.
a, 3D rendering of the bottom third of a live, Lifeact–eGFP-expressing metaphase HeLa imaged by SDCM. The cortical actin signal is well resolved, whereas lower-contrast deep subcortical actin assemblies appear hazy and ill-defined. b, Shadow projection of subcortical and cytoplasmic actin in a fixed, phalloidin–AF488-stained HeLa cell. Expanded region shows 3D meshwork of actin cables. Volume was acquired by Airyscan microscopy and the brighter cortical actin signal was computationally masked and removed. c, Blend projection of actin cables (AF488–phalloidin) in a metaphase HeLa cell. Volume was acquired by Airyscan and processed as in b. d, z-coded maximum-intensity projection of the actin meshwork (AF488–phalloidin) in a prometaphase HeLa with expanded region. e, Manual segmentation of actin cables in one half of a metaphase HeLa cell. f, Single-slice, Airyscan image of metaphase actin cables (AF488–phalloidin). Segmented cables are pseudocoloured by relative orientation (−90° to 90°). g, Cartoon schematic of actin cable meshwork indicating parameters used for analysis. h, Representative actin cable analysis workflow showing ridge detection to identify and skeletonize cables, Euclidean distance mapping to identify pore centres, and largest circle fitting technique to estimate maximum pore size. All analyses were performed on 2D slices in the basal metaphase cytoplasm. i, Estimated width of actin cables and 100-nm TetraSpeck beads imaged by GI-SIM. Widths are based on full width at half maximum of Gaussian fits of 1-pixel line scans through individual cables. j, Frequency distribution of actin mesh pore radii. k, Area of largest actin meshwork fitting circle. l, Actin cable density in metaphase or anaphase HeLa cells. m, Airyscan image of actin cables (phalloidin), DNA (Hoechst) and centrosomes (anti-pericentrin) in a central slice of an anaphase HeLa cell. A magnified view of actin cables in the basal cytoplasm is shown on the right. n, GI-SIM image of mitochondria (mito–dsRed2) co-orientation with actin cables (Lifeact–eGFP). o, Airyscan images of mitochondria associated with actin cables in live metaphase HeLa cells. p, Mitochondria (anti-TOM20) associated with actin cables (phalloidin) in a metaphase U2OS cell. q, Mitochondria (anti-TOM20) associated with actin cables (phalloidin) in a metaphase HEK 293T cell. r, Airyscan montage and cartoon of a mitochondrion oscillating between actin cables in the metaphase cytoplasm. s, Left, raw Airyscan image of mitochondria (mito–dsRed2) and actin cables (Lifeact–eGFP, arrows) in the deep cytoplasm of a metaphase HeLa cell. Right, segmented, median-time averaged image of the same region used in Fig. 1d. Kymograph indicating an individual mitochondrion oscillating between two neighbouring actin cables. t, Representative 44-s trajectory of mitochondria within the actin cable mesh. u, Cumulative squared displacement of mitochondria diffusing within the actin cable meshwork. v, Montage and kymograph of transient mitochondrial co-motility with a local actin cable. w, Cartoon indicating co-motility between a cable and an immediately adjacent (local) mitochondrion. Instantaneous velocity correlation coefficient for actin cables with either local or distal mitochondria. Scale bars: 10 μm (a, c, e, m, left, p); 5 μm (b, d, q); 1 μm (b inset, d inset, f, h, n, o, p inset, r, s, t, v); 2.5 μm (m right, q inset). Sample sizes: 100 cables from 20 cells (i); 28 cells (j, k); 46 metaphase cells, 25 anaphase cells (l); 19 cells (w). Samples were drawn from at least three independent experiments. Statistical tests: two-tailed unpaired t-test ***P < 0.0001 (l, w); median ± interquartile range (i–l, w).
Extended Data Fig. 3 Disruption of the actin cable meshwork impairs metaphase mitochondrial motility and distribution.
a, F-actin (Lifeact–eGFP) and mitochondria (mito–dsRed2) organization in untreated, LatA-treated (1 μM, 1 h) and CytoD-treated (100 nM, 1 h) metaphase HeLa cells. Small arrows indicate actin cable network in untreated cells. Large arrows indicate aggregated actin cables in CytoD-treated cell. b, Metaphase HeLa treated with 1 μM LatA for 1 h. LatA treatment eliminated subcortical actin structures (Lifeact–eGFP) with minimal effects on cell geometry. White arrow indicates region of clumped mitochondria (mito–dsRed2). Yellow arrows indicate cytoplasmic regions with decreased mitochondrial density. c, Representative metaphase HeLa cells treated with 100 nM CytoD for 1 h. CytoD treatment induced collapse of the meshwork (Lifeact–eGFP) and marked disorganization of actin cables. As with LatA treatment, CytoD treatment induced mitochondrial aggregation (mito–dsRed2). d, MSD of 60-s mitochondrial trajectories in untreated, LatA-treated or CytoD-treated metaphase cells. e, Velocity autocorrelation at 0.5-s delays (Cv(0.5 s)) for mitochondrial trajectories from untreated, LatA-treated or CytoD-treated metaphase HeLa cells. f, Discretized velocity autocorrelations of 60-s mitochondrial trajectories in untreated, CytoD-treated or LatA-treated cells. g, Left, 3D segmentation mask of mitochondria (mito–dsRed2) in a live metaphase HeLa cell (pseudocoloured by z-position). Right, eroded points derived from the mitochondrial mask with schematic indicating spherical coordinate system with origin at the cell centroid. h, Schematic indicating 3D mitochondrial distribution analyses. i, 3D mitochondrial mean centre distance to cell centroid in untreated or CytoD-treated cells. j, Standard deviation of the percentage of the mitochondrial network in each of 6 equal sized 3D wedges from control (untreated) or CytoD-treated cells. Wedges were specified by azimuthal values of 0°–120°, 120°–240° and 240°–360° in either the bottom hemisphere (that is, elevation −90° to 0°) or the top hemisphere (that is, elevation 0° to 90°). k, 3D resultant vector length for control (untreated) or CytoD-treated cells. l, Scatter plot of nucleoid radial (y axis) and angular (x axis) positions superimposed on histograms of nucleoid count per 60° from DMSO- and CytoD-treated cells in Fig. 1g. m, Schematic indicating 2D mtDNA nucleoid distribution analyses. n, Example rotational asymmetry traces of nucleoids in a DMSO- or LatA-treated metaphase cell. Asymmetry was calculated as the ratio of nucleoids on side A to the total number of nucleoids (sides A + B) upon rotating the dividing chord by 1° intervals over 360°. o, Variance of rotational asymmetry over 360° for DMSO-, CytoD- or LatA-treated cells. p, Schematic indicating Euclidean and circular nearest-neighbour distance analysis for nucleoids. Index of dispersion for ten nearest neighbours of all nucleoids in a cell is reported. q, Index of dispersion of Euclidean nearest-neighbour distances between all nucleoids and their ten nearest neighbours in cells treated with DMSO, LatA or CytoD. r, Index of dispersion of circular nearest neighbour distances between all nucleoids and their ten nearest neighbours in cells treated with DMSO, LatA or CytoD. s, Mean nucleoid radial position in cells treated with DMSO, CytoD or LatA. t, Mitochondrial nucleoids (PicoGreen) and plasma membrane (Cell Mask Orange) in metaphase A549 cells treated with DMSO (left) or 100 nM CytoD (right). u, Mitochondrial nucleoids (PicoGreen) and plasma membrane (Cell Mask Orange) in metaphase HEK 293T cells treated with DMSO (left) or 100 nM CytoD (right). v, Mitochondrial nucleoids (PicoGreen) and plasma membrane (Cell Mask Orange) in metaphase iPS cells treated with DMSO (left) or 100 nM CytoD (right). Scale bars: 2.5 μm (a); 10 μm (b, c, g, t–v). Sample sizes: 479 control, 232 LatA-treated and 185 CytoD-treated mitochondria trajectories (d–f); 26 untreated, 14 CytoD-treated cells (i–k); 36 DMSO-treated, 33 LatA-treated and 35 Cyto-D-treated cells (o, q–s). Samples were drawn from at least three independent experiments. Statistical tests: Kruskal–Wallis test with Dunn’s multiple comparisons test ***P < 0.0001 (e, o); two-tailed Mann–Whitney test **P = 0.0099 (i); two-tailed unpaired t-test ***P < 0.0001 (j); two-tailed unpaired t-test **P = 0.0025 (k); two-way repeated measures ANOVA with Dunnet’s multiple comparisons test (q), neighbour 1: DMSO vs LatA P = 0.0833, DMSO vs CytoD *P = 0.0288; neighbour 2: DMSO vs LatA ***P < 0.0001, DMSO vs CytoD ***P = 0.0002; neighbour 3: DMSO vs LatA ***P < 0.0001, DMSO vs CytoD **P = 0.0026; neighbour 4: DMSO vs LatA ***P < 0.0001, DMSO vs CytoD ***P = 0.0004; neighbour 5: DMSO vs LatA ***P < 0.0001, DMSO vs CytoD ***P = 0.0002; neighbours 6–10: DMSO vs LatA ***P < 0.0001, DMSO vs CytoD ***P < 0.0001. Kruskal–Wallis test with Dunn’s multiple comparisons test (s), DMSO vs LatA P = 0.2294, DMSO vs CytoD P = 0.6893. Median ± interquartile range (e, i–k, o, s); mean ± s.e.m. (q–r).
Extended Data Fig. 4 Disruption of the actin cable meshwork results in metaphase ER collapse.
a, iSIM slice of ER (eGFP–Sec61B) and mitochondria (MitoTracker DeepRed) in a prometaphase HeLa. b, iSIM slice of ER (eGFP–Sec61B) and mitochondria (MitoTracker DeepRed) in a metaphase HeLa. c, iSIM image of ER (eGFP–Sec61B), mitochondria (MitoTracker DeepRed) and actin (Lifeact–mScarlet) with accompanying segmented and masked images. d, Airyscan image of mito–dsRed2 (blue), Halo–Sec61B (grey) and Lifeact–eGFP (orange) in a metaphase HeLa cell with accompanying masked images of actin cables and ER profiles. e, Airyscan montage of ER cisternal collapse after addition of 1 μM LatA. f, STED image of ER (Halo–KDEL) in a metaphase HeLa treated with 100 nM CytoD for 1 h. Arrows indicate tightly stacked ER sheets. Asterisk indicates expanded cytoplasmic void between ER sheets. g, Representative analysis workflow for estimating the size of voids between ER sheets. h, Time points at 0 and 12 min from e are shown with void centre positions in orange. i, Index of dispersion (variance-to-mean ratio) of areas of cytoplasmic voids from the cell shown in e over 24 min. LatA (1 μM) was added at time 0. j–m, Inter-cisternal ER void size analyses in DMSO- or CytoD-treated cells, including mean inter-cisternal void area (j), maximum void area (k), standard deviation of void area (l) and index of dispersion of void area (m). n–q, Inter-cisternal ER void centre position analyses, including void mean centre distance to cell centroid (n), void centre resultant vector length (o), s.d. of void centre occupancy per 60° sector (p) and void centre radial position (q). r, iSIM image of Lifeact–mScarlet, eGFP–Sec61B and MTDR in a metaphase HeLa treated with CytoD for 1 h. Magnified inset is shown in Fig. 1l. s, Representative example of cisternal collapse and mitochondrial confinement in a metaphase HeLa treated with LatA (1 μM, 1.5 h). t, Airyscan image of the cell from e at 20 min showing both ER (Halo–Sec61B) and mitochondria (mito–dsRed2). u, Mean distance from void centre positions to nearest mitochondria is significantly increased in CytoD-treated cells. v, Cartoon indicating close association between mitochondria (blue) and both ER sheets (grey) and actin cables (orange). Elimination of actin cables by LatA or CytoD results in ER disorganization, characterized by inter-cisternal ER void collapse and expansion, effectively displacing mitochondria from regions of the metaphase cytoplasm. Scale bars: 10 μm (a, b, e, h, r–t); 2.5 μm (a inset, b inset, g, s inset); 5 μm (c, d, f). Sample sizes: 22 DMSO- and 31 CytoD-treated cells (j–q); 16 DMSO-treated, 14 CytoD-treated cells (u). Samples were drawn from at least three independent experiments. Statistical tests: two-tailed unpaired t-test ***P < 0.0001 (j, l, n); two-tailed unpaired t-test ***P = 0.0003 (k); two-tailed Mann–Whitney test ***P < 0.0001 (m, u); two-tailed unpaired t-test, **P = 0.0016 (o); two-tailed unpaired t-test, ***P = 0.0002 (p); two-tailed unpaired t-test, P = 0.0975 (q). Median ± interquartile range (j–q, u).
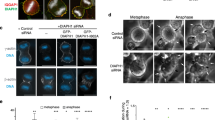
Extended Data Fig. 5 Myosin19 links mitochondria and associated ER membranes to the actin cable meshwork.
a, Immunoblot of MYO19 in HeLa cells treated with nontargeting (NT) siRNA or MYO19 siRNA (right). Relative total protein levels were used as a loading control. Knockdown efficiency was 96%, based on 3 independent experiments. b, Spinning disk image of mitochondria (mito–dsRed2) and DNA (Hoechst) in fixed metaphase Hela cells treated with NT or MYO19 siRNA. Arrows indicate regions of metaphase cytoplasm devoid of mitochondria. c, Schematic indicating parameters used for analyses in d and e, including xy centre of mitochondrial mass, xy cell centroid, and 60° circle sectors. d, Centre of mitochondrial mass displacement from cell centroid in metaphase HeLa cells treated with DMSO (1 h), CytoD (100 nM, 1 h), NT siRNA (40 nM, 48 h) or MYO19 siRNA (40 nM, 48 h). e, Standard deviation of mitochondrial mass per 60° circle sector in metaphase HeLa cells treated with DMSO (1 h), CytoD (100 nM, 1 h), NT siRNA (40 nM, 48 h), or MYO19 siRNA (40 nM, 48 h). f, 3D rendering of mitochondria (mito–dsRed2) and DNA (Hoechst) in cytokinetic HeLa cells treated with NT siRNA. g, 3D rendering of mitochondria (mito–dsRed2) and DNA (Hoechst) in cytokinetic HeLa cells treated with MYO19 siRNA. h, Inheritance index (A/B) of mitochondria in cytokinetic HeLa cells treated with either NT or MYO19 siRNA. Ratio is defined as mitochondrial mass in daughter cell A/mitochondrial mass in daughter cell B, where A is the daughter cell with fewer mitochondria. i, Mitochondria (mito–dsRed2) and actin cables (Lifeact-eGFP) in metaphase HeLa cells treated with NT or MYO19 siRNA. Bottom row, segmented images of mitochondria and actin cables. White circle indicates a region of the cable meshwork largely devoid of mitochondria. j, Cartoon of mitochondria and actin cables in the cytoplasm of NT siRNA- or MYO19 siRNA-treated metaphase cells. k, Mean distance (μm) from actin cables to the nearest mitochondrial pixel. Median ± interquartile range. l, m, Airyscan images of mitochondria (mito–dsRed2) and ER cisternae (Halo–Sec61) in metaphase HeLa cells treated with NT siRNA (l) or MYO19 siRNA (m) for 48 h. Arrows indicate collapsed, compacted ER sheets. Asterisks indicate expanded, inter-sheet cytoplasmic voids. Scale bars: 10 μm (b, f, g, i, l, m); 5 μm (l inset, m inset). Sample sizes: 17 DMSO-, 28 CytoD-, 44 NT siRNA- and 19 MYO19 siRNA-treated cells (d, e); 29 NT siRNA-, 33 MYO19 siRNA-treated cells (h); 12 NT siRNA-, 13 MYO19 siRNA-treated cells (k). Samples were drawn from at least three independent experiments. Statistical tests: Ordinary one-way ANOVA with Tukey’s multiple comparisons test, DMSO vs CytoD **P = 0.0087, CytoD vs NT **P = 0.0011, NT vs MYO19 **P = 0.0012, DMSO vs MYO19 **P = 0.006, DMSO vs NT P = 0.9962, CytoD vs MYO19 P = 0.975 (d); ordinary one-way ANOVA with Tukey’s multiple comparisons test ****P < 0.0001, DMSO vs NT P = 0.9733, CytoD vs MYO19 P = 0.913 (e); two-tailed Mann–Whitney test ***P < 0.0001 (h); two-tailed unpaired t-test ***P < 0.0001 (k). Median ± interquartile range (d, e, h, k). An uncropped and unprocessed scan of the blots in a is included in the Source Data.
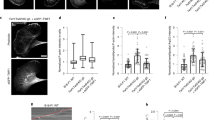
Extended Data Fig. 6 Mitochondrial actin waves rapidly cycle through mitotic mitochondrial networks.
a, Top, spinning disk confocal montage of mitochondria (MitoTrackerDeepRed, MTDR) and F-actin (Lifeact–mScarlet). Bottom, monochrome montage of Lifeact signal with yellow arrow indicating position of the mitochondrial actin wave. Scale bar, 10 μm. b, 3D kymograph of metaphase actin wave upon subtraction of cortical actin signal. c, Interphase HeLa cell expressing Lifeact–eGFP and mito–dsRed2. Insets indicate mitochondria inside or outside of the actin wave (arrows). d, Autocorrelation of interphase actin wave. Mean ± 95% confidence interval. e, Directionality of actin waves. Negative values on the x axis indicate a net anticlockwise bias after 100 min, while positive values indicate a net clockwise bias. f, Fraction of HeLa cells in the indicated stages of mitosis with mitochondrial actin waves. g, Table comparing mitochondrial actin waves described here and previously described revolving actin clusters12. h, Representative images of F-actin (Lifeact–eGFP) and mitochondria (mito–dsRed2) in HeLa cells in different stages of mitosis. White arrows indicate position of the actin wave. i, Airyscan image of F-actin (phalloidin) and peroxisomes (eGFP–SKL) in a fixed, metaphase HeLa cell. Dashed line indicates boundary of the actin wave. Arrows indicate peroxisomes outside of actin clouds. j, Airyscan images of F-actin (Lifeact) and late endosomes (eGFP–Rab7a) in a live, metaphase HeLa cell. Dashed line indicates boundary of the actin wave. Arrows indicate endosomes outside of actin clouds. k, Airyscan images of F-actin (Lifeact) and lysosomes (SiR-lysosome) in a live, metaphase HeLa cell. Dashed line indicates boundary of the actin wave. Arrows indicate lysosomes outside of actin clouds. l, Airyscan images of F-actin (phalloidin) and ER (Halo–Sec61) in a fixed, metaphase HeLa cell. Dashed line indicates boundary of the actin wave. Arrows indicate ER outside of actin clouds. Scale bars: 10 μm (a, c, h–l); 2.5 μm (b, centre, i–l, insets); 1 μm (c, right). Sample sizes: 17 cells (d), 10 cells (e), 167 cells (f). Samples were drawn from at least three independent experiments.
Extended Data Fig. 7 Mitochondria associate with actin cables, clouds, and comet tails in mitosis.
a, Gamma-adjusted version of the image shown in Fig. 2h, revealing lower-intensity actin cables outside of the wave. Insets show mitochondria outside of the wave associated with actin cables. b, Live metaphase HeLa cell expressing Lifeact-eGFP and mito–dsRed2 with insets indicating position of actin wave and cables. c. Airyscan image of mitochondrial actin clouds within a metaphase actin wave. White dashed line indicates wave border. Expanded regions show mitochondria associated with cables (arrows) outside of the wave. d, Relative phalloidin intensity of actin clouds and actin cables. Values represent the ratio of clouds or cables to cortical actin signal. e, Representative Airyscan images of mitochondrial actin clouds with clipped images showing bright actin nodes on the surface of mitochondria. f, Representative kymographs indicating restricted mitochondrial motility inside of actin clouds. g, Airyscan montage of an actin cloud symmetry breaking event. Arrows indicates a gap forming between adjacent nodes in the cloud preceding mitochondrial ejection. h, i, Representative images of mitochondria associated with actin comet tails in live HeLa cells. j. Airyscan maximum-intensity projection of a mitochondrial actin comet tail in a metaphase HeLa. k, Metaphase mitochondrial actin comet tails from the indicated cell types. Arrows indicate trailing comet tails. l, Speed of comet tail-based motility in the indicated cell types. m, Airyscan image of actin (Lifeact–eGFP) and mitochondria (mito–dsRed2) in a metaphase HeLa cell. Montage indicates motility of mitochondria associated with a cloud (top) or comet tail (bottom) over 39 s. n, Airyscan montage of mitochondrial comet tail extension. o, Spinning disk montage of F-actin (Lifeact–eGFP) and mitochondria (mito–dsRed2) upon comet tail assembly and extension. p, Montage of actin comet tail regaining symmetry, forming an actin cloud around the immotile mitochondrion. q, Montage of a mitochondrion (yellow arrow) undergoing comet tail motility resulting in movement out of the imaging plane. r, xz projection of F-actin (phalloidin), mitochondria (mito–dsRed2), and DNA (Hoechst) in a fixed, metaphase HeLa cell. Inset indicates a mitochondrion associated with an actin comet tail that is oriented primarily in the axial plane. s, 3D trajectory of a comet tail-propelled mitochondrion from a 3D Airyscan-fast video. Trajectory is coloured by time. t, Actin comet tail parameters estimated from 3D volumes of fixed, phalloidin stained cells. u, Mean speed of mitochondria associated with the indicated actin structures over 25 s. v, Density estimation of instantaneous speeds of cable, cloud and comet tail associated mitochondria. w, Speed of comet tails in metaphase HeLa treated with DMSO or 25 μM nocodazole. x, Three-minute time projection of mitochondria (mito–dsRed2) in a MYO19 siRNA-treated metaphase HeLa cell. Comet tail trajectories are shown in yellow and in the expanded inset. Asterisk indicates regions of cytoplasm unexplored by mitochondria over 3 min. y, MSD of cable-, cloud- and comet-associated mitochondria. MSD plot with cropped y axis is shown on the right to visualize the reduced cable and cloud motility. z, Velocity autocorrelations of the three types of motility. Cv(0.5 s) values are plotted for mitochondria associated with cables, clouds, comet tails, as well as mitochondria in fixed cells to estimate tracking error. Scale bars: 10 μm (a–c, m, r, x); 2.5 μm (a, insets, b, insets, r, inset); 1 μm (c, inset, e–k, m, right, n–q). Sample size: 16 cells (d); 233 HeLa, 40 Cos7, 35 A549, 30 HaCaT, 74 HEK 293T (l); 59 cables, 59 clouds, 47 comets (u, v); 38 DMSO, 61 nocodazole-treated cells (w). 57 cables, 47 comets and 59 clouds (y, z). Samples were drawn from at least three independent experiments. Statistics: two-tailed unpaired t-test ***P < 0.0001 (d); Kruskal–Wallis test with Dunn’s multiple comparisons test ***P < 0.0001 (u); two-tailed Mann–Whitney test *P = 0.011 (w); ordinary one-way ANOVA with Tukey’s multiple comparisons test ***P < 0.0001 (z). Median (d); mean ± s.d. (l, t); median ± interquartile range (u, w, z, right); mean ± 95% confidence interval (y, z, left).
Extended Data Fig. 8 Mitochondrial actin waves are maintained by multiple actin assembly and disassembly factors.
a, Mitochondrial actin wave area in cells treated with indicated inhibitors. b, Table indicating the drugs used in a, as well as their major targets and concentrations and incubation times used. c, Mitochondrial actin wave area in cells treated with indicated siRNA for 48 h. d, Knockdown efficiency of siRNAs used in c. Values indicate averages of at least three independents experiments. e, Representative western blots indicating knockdown efficiency of siRNAs in c. Full blots are shown in Supplementary Fig. 1. Relative total protein levels are indicated below each blot. f, Representative single-plane images of actin waves (Lifeact–eGFP) in metaphase HeLa cells treated with the indicated siRNAs. Dashed red line indicates wave boundaries. g, Airyscan single plane image of a HeLa cell expressing eGFP–VASP (white), Lifeact–mScarlet, (red) and Mito–TagBFP2 (blue). Yellow arrows indicate position of VASP puncta on the actin cloud. h, Airyscan image of VASP association with a mitochondrion (Mito–TagBFP2) within an actin cloud (Lifeact–mScarlet). Right, normalized fluorescence intensity of Mito, VASP and Lifeact along the 1-pixel linescan indicated by the dashed line. i, Metaphase HeLa cells expressing Lifeact–mScarlet, filamin A–eGFP and MitoTracker DeepRed. j, Fixed, anaphase HeLa cell stained for filamin A (cyan) and F-actin (phalloidin, orange). k, Spinning disk confocal median time projection (over 30 s) indicating cortactin–eGFP colocalization with Lifeact–mScarlet in a metaphase actin wave. l, Median intensity time projection of Lifeact–mScarlet and cortactin–eGFP colocalization in an actin cloud in a metaphase HeLa cell. m, SDCM images of fixed HeLa cells stained for mitochondria (anti-TOM20), cortactin (anti-cortactin) and F-actin (phalloidin). White arrows indicate asymmetric, punctate localization of cortactin around actin-positive mitochondria. Right, normalized fluorescence intensity of F-actin (phalloidin), TOM20 and cortactin along the 1-pixel-wide line scan. n, Colocalization of α-actinin (α-actinin–mNeonGreen) and F-actin (Life act–mScarlet) in a metaphase HeLa cell. Right, kymograph of LifeAct and α-actinin generated from the indicated line scan (dashed line). Scale bars: 10 μm (f, i–k, n); 5 μm (g, l); 1 μm (e, g, inset, l, inset, m). Sample sizes: number of cells per condition is indicated above each box plot (a, c). Samples are drawn from at least three independent experiments. Statistical tests: Kruskal–Wallis test with Dunn’s multiple comparisons test ***P < 0.0001, DMSO vs C3 Transferase P = 0.7515, DMSO vs rhosin P < 0.9999 (a); Kruskal–Wallis test with Dunn’s multiple comparisons test ***P < 0.0001, NT vs WASH1 P > 0.9999, NT vs WAVE1 P = 0.6079, NT vs WHAMM P = 0.0766, NT vs N-WASP (also known as WASL) P > 0.9999, NT vs MYO19 P > 0.9999, NT vs MYO5A P > 0.9999 (b). In box plots, centre line shows median, box encompasses the interquartile range, whiskers span 10th to 90th percentiles and ‘+’ indicates the mean. Uncropped and unprocessed scans of the blots are included in the Source Data.
Extended Data Fig. 9 ARP3 depletion eliminates actin comet tail-based mitochondrial motility but does not impair mitochondrial positioning or equal inheritance.
a, Frequency of mitochondrial comet tail-based motility over 1 min in metaphase Hela cells treated with NT or ARP3 siRNA. Median ± interquartile range. n = 11 NT siRNA-treated cells, 11 ARP3 siRNA-treated cells from at least three independent experiments. b, Left, three-frame median-intensity time projection of actin cables (Lifeact–eGFP) and mitochondria (mito–dsRed2) in an ARP3-depleted metaphase HeLa cell. Right, time-coded maximum-intensity projection of segmented actin cables over 178 s. c, Airyscan image of mitochondria (mito–dsRed2) associated with actin cables (Lifeact–eGFP) in an ARP3 siRNA-treated metaphase HeLa cell. Kymograph shows mitochondria and cable motility over ~2 min. d, Single-plane Airyscan image of mitochondria (mito–dsRed2) in an NT siRNA-treated (left) or ARP3 siRNA-treated (right) metaphase HeLa cell. e, Centre of mitochondrial mass displacement from cell centroid in cells treated with NT or ARP3 siRNA. Note: the NT data are reproduced from Extended Data Fig. 5e. n = 44 NT, 38 ARP3 from at least three independent experiments. f, Standard deviation of mitochondrial per cent occupancy per 60° sector in cells treated with NT or ARP3 siRNA. g, Origin-aligned trajectories for 150 mitochondria over 60 s. Control trajectories were taken from outside of the actin wave in untreated cells. ARP3 siRNA trajectories are taken from random positions in the cell. h, Single-plane cumulative maximum-intensity projections of mitochondria (mito–dsRed2) in NT or ARP3 siRNA-treated metaphase HeLa cells. i, Percentage of cell area covered by mitochondria per min in NT or ARP3 siRNA-treated metaphase HeLa cells. j, Displacement index of mitochondria in NT or ARP3 siRNA-treated HeLa cells. Displacement index is calculated by dividing mito–dsRed2 area in 5-min maximum intensity time projections by the area of mito–dsRed2 in the first frame. k, Mitochondria (mito–dsRed2) and ER (Halo–SEC61) in an ARP3 siRNA-treated HeLa cell. l, Equal mitochondrial inheritance in an ARP3 siRNA-treated HeLa cell undergoing cytokinesis. Dashed line indicates division plane. m, Inheritance ratio (A/B) of metaphase HeLa cells treated with NT or ARP3 siRNA. Note: NT siRNA data are reproduced from Extended Data Fig. 5. The ratio is defined as mitochondrial mass in daughter cell A/ mitochondrial mass in daughter cell B, where A is the daughter cell with fewer mitochondria. n, Change in distance between pairs of neighbouring mitochondria after 5 min in a NT or ARP3 siRNA-treated metaphase HeLa cell. Scale bars: 1 μm (b); 2.5 μm (c, k, inset); 5 μm (d); 500 nm (g); 10 μm (h, k, l). Sample sizes: the NT data are reproduced from Extended Data Fig. 5e, 44 NT, 38 ARP3 siRNA-treated cells from at least three independent experiments (e); the NT data are reproduced from Extended Data Fig. 5e, 44 NT, 38 ARP3 siRNA-treated cells from at least three independent experiments (f); 38 NT, 33 ARP3 siRNA-treated cells (i, j); 29 NT siRNA, 14 ARP3 siRNA-treated cells (m); change in neighbour distance was calculated on one NT siRNA and one ARP3 siRNA-treated cell (n). Statistical tests: two-tailed unpaired t-test, ***P < 0.0001 (i, j). Median ± interquartile range (a, e, f, i, j, m).
Extended Data Fig. 10 Actin waves shuffle mitochondrial position before cytokinesis.
a, b, Airyscan maximum-intensity projections of NT siRNA-treated (a) or ARP3 siRNA-treated (b) metaphase HeLa cells expressing mito–paGFP and mito–KR. White box indicates region irradiated with 405-nm and 561-nm light to simultaneously bleach Mito–KR and activate mito–paGFP. c, Computationally straightened montage of mito–paGFP dispersion in NT or ARP3 siRNA-treated cells d, Mean rotational asymmetry of damaged mitochondria in NT or ARP3 siRNA-treated cells at 0 and 20 min after photoactivation. e, Displacement of damaged, mito–paGFP-labelled mitochondria centre of mass per min in NT or ARP3 siRNA-treated cells. f, Spinning disk maximum-intensity projection of mito–paGFP and mito–KR in metaphase HeLa cells treated with DMSO (left) or CK-666 (right). White boxes indicate regions irradiated with 405-nm and 561-nm light to simultaneously bleach Mito–KR and activate mito–paGFP. g, Airyscan maximum-intensity projection of an ARP3 siRNA-treated metaphase HeLa cell expressing mito–paGFP and Lifeact–mScarlet. White box indicates region irradiated with 405-nm light to activate mito–paGFP. h, Circular kernel density estimate of mito–paGFP position over 40 min. Dashed line indicates midpoint of the photoactivation region at time 0. i, Airyscan image of MFN1 and MFN2 siRNA (MFN1/MFN2)-treated interphase HeLa cell expressing Lifeact–eGFP (orange) and mito–dsRed2 (blue). Dashed line indicates position of the actin wave. Expanded view shows actin-positive mitochondria within the wave. j, Airyscan image of fragmented, morphologically simple mitochondria in interphase HeLa cells treated with MFN1 and MFN2 siRNA. k, Metaphase actin wave (black arrow) persist upon simultaneous depletion of the mitochondrial fusion factors MFN1 and MFN2. l, Representative example of actin comet tail-based mitochondrial movements in a metaphase MFN1 and MFN2 siRNA-treated HeLa. m, Airyscan maximum-intensity projections of a MFN1 and MFN2 siRNA-treated metaphase HeLa cell expressing Mito-Dendra2. White box indicates region irradiated with 405-nm light to photoconvert the Dendra2. Over 21 min the photoactivated mitochondria are dispersed through the cell. n, Circular kernel density estimates of Mito–Dendra2 distribution over 21 min, indicating increased dispersion of the photoconverted mitochondria over time. o, Histogram indicating simulated damaged mitochondrial dispersion over 30 min. Grey boxes indicate net mitochondrial angle change in simulations with actin waves, red boxes indicate net angle change in simulations without waves. p, Estimated cumulative density of damaged mitochondria angle change over 30 min in simulations with different sized actin waves. q, Estimated cumulative density of damaged mitochondria angle change over 30 min in simulations with different speed actin comet tails. r, s, Percentage of damaged (orange) and total (grey) mitochondria inherited by daughter cell B over 30 min in simulations with variable wave sizes or actin comet tail speed. Mean ± 95% confidence interval. Scale bars: 10 μm (a, b, f, g, i, m); 5 μm (i, inset, j); 1 μm (l). Sample sizes: 13 NT siRNA-, 14 ARP3 siRNA-treated cells (d, e). Samples are drawn from at least three independent experiments. Statistical tests: two-way repeated measures ANOVA with Sidak’s multiple comparisons test (d), NT vs ARP3 at 0 min, P = 0.9927, NT vs ARP3 at 20 min, ***P = 0.0006; two-tailed unpaired t-test, **P = 0.0013 (e). Median ± interquartile range (e).
Supplementary information
Supplementary Figure 1
Probed blots and total protein levels are shown for western blots confirming knockdown of Myo19 (Extended Data Fig. 5a), Arp3 (Extended Data Fig. 8e), WASH1 (Extended Data Fig. 8e), WAVE1 (Extended Data Fig. 8e), WHAMM (Extended Data Fig. 8e), N-WASP (Extended Data Fig. 8e), VASP (Extended Data Fig. 8e), INF2 (Extended Data Fig. 8e), INF2-CAAX (Extended Data Fig. 8e), Cofilin (Extended Data Fig. 8e), Gelsolin (Extended Data Fig. 8e), ADF (Extended Data Fig. 8e), and MyoVa (Extended Data Fig. 8e). Total protein stains were measured from the same lanes. NT = control nontargeting siRNA. Red box indicates cropped regions for figure. Asterisks indicates that the cropped region was flipped horizontally for clearer display in the extended data figure.
Video 1
Mitochondria motility within the metaphase actin cable meshwork. a. Airyscan maximum intensity projection (5 slices, 4 µm deep) time-lapse of mitochondria (blue) and actin cables (orange) in metaphase cytoplasm. b. Airyscan single plane time-lapse of mitochondria (blue) and actin cables (orange) in a metaphase HeLa. Time averaged time-lapse of segmented mitochondria and cables is also shown. c. Airyscan time-lapse of mitochondria (, magenta) and actin cables (green) at 0.5 sec per frame. Trajectories of each structures are shown over 31 sec. d. Additional examples of mitochondria and actin cable motility in 2D airyscan time-lapses. Scale barss = 1 µm. Time = mm:ss.
Video 2
Metaphase ER collapse after actin cable disassembly. Airyscan time-lapse of actin cables (Lifeact-EGFP, green) and ER membrane (Halo-SEC61B, magenta) in metaphase arrested HeLa cells (10 µM nocodazole). Scale = 2 µm. Time = mm:ss. Next, an Airyscan time-lapse of mitochondria (mito–dsRed2, blue) and ER membrane (Halo-SEC61B) in a metaphase HeLa treated with 1 µM latrunculin A at time 0. Comparison of 00:00 and 23:55 is shown at the end. Scale bar = 5 µm.
Video 3
Mitochondrial actin waves persist throughout cell division. a. Actin waves cycling through interphase mitochondrial networks. Time = hh:mm:ss. Playback speed increases at 01:04:00. b. Actin waves (arrows) cycling through interphase mitochondrial networks. Cell on the right enters mitosis at ~200:00. Time=mmm:ss. c. Actin wave (arrow) cycling through a metaphase HeLa mitochondrial network. Time = mm:ss. d. Mitochondrial actin wave (arrows) in an anaphase HeLa. Time = mm:ss. e. Actin wave stalling on mitochondria localized to the cleavage furrow. Time = mm:ss. f. Mitochondrial actin waves in a population of interphase HeLa acquired at 90 sec per frame. Note the actin wave in the cell on the right that persists after completion of mitosis. time = hh:mm:ss.
Video 4
Mitochondrial actin waves in multiple metaphase cell types. a. Mitochondrial actin wave in a metaphase HeLa cell. Time = mm:ss. Next, a surface plot of Lifeact-mScarlet is shown after manually removing cortical actin signal. Next, additional examples of metaphase HeLa actin waves (Lifeact-EGFP, black) are shown to demonstrate different cycling directions. Finally, actin accumulation on mitochondrial outer membranes in a metaphase HeLa is shown by multiplying binary masks of the mitochondrial OMM with the lifeact signal. Note the persistent clockwise wave. B. Additional examples of actin waves (Lifeact) cycling through metaphase mitochondrial networks in normal human epidermal keratinocytes (NHEK), HaCaT, HEK 293T, COS-7, and iPS cells.
Video 5
3D actin waves in the metaphase cytoplasm. a. Airyscan 3D rendering of subcortical actin structures (Phalloidin) in the bottom half of a fixed metaphase HeLa cell. The video pans in on an actin wave adjacent to the actin cable meshwork (arrows). B. Lattice light-sheet 3D rendering of a metaphase HeLa cell with a counter-clockwise cycling actin wave (Lifeact-EGFP) revealed to be cycling through a mitochondrial network (Mito-dsRed2) at ~ 17:25. Time = mm:ss. C. 3D segmentation of mitochondrial actin wave from the cell in B.
Video 6
Spontaneous mitochondrial cloud-to-comet transitions. A. Mitochondrial actin cloud symmetry breaking followed by the propulsion of mitochondria by actin comet tails. Mitochondria = Mito-dsRed2, blue. F-actin = Lifeact-EGFP, orange. Scale = 1 µm. Time = mm:ss. B. Spinning disk time-lapse of a mitochondrial cloud converting to a comet tail (Mito-dsRed2, red; Lifeact-EGFP, white). Time = mm:ss. Scale = 2 µm.
Video 7
Actin comet tails drive directed bursts of mitochondrial motility. A. Actin comet tail based mitochondrial motility in metaphase HeLa cells. Time = mm:ss. Scale bar = 1 µm. B. Computationally stabilized mitochondrial comet tail. Scale bar = 0.5 µm, time = mm:ss. C. Computationally straightened or “linearized” comet tail trajectory. Scale bar = 1 µm, time = mm:ss. D. Spinning disk time-lapse of a metaphase HeLa cells expressing lifeact-EGFP (white) and mito-dsRed2 (orange). Mitochondrial comet trajectories are shown (right) over 05:58. Time = mm:ss. Scale bar = 10 µm.
Video 8
Mitochondria motility within the actin cable meshwork in an Arp3 siRNA-treated metaphase HeLa. Airyscan time-lapse of mitochondria (Mito-dsRed2, blue) and F-actin (Lifeact-EGFP, orange) in an Arp3 siRNA-treated metaphase HeLa cell. Inset shows mitochondrial motility within the cable meshwork. To more easily visualize actin cables, a median time projection was applied over three frame windows. Scale = 10 µm, time = mm:ss.
Video 9
Actin waves disperse clusters of damaged mitochondria. a-e. Airyscan maximum intensity projection time-lapses of NT siRNA- or Arp3 siRNA-treated HeLa cells expressing mito-paGFP and mito-KillerRed from Fig. 3a (a-c) or Extended Data Fig. 10a-b (d-e). f. Airyscan maximum intensity projection time-lapse of MFN1/MFN2 siRNA-treated metaphase HeLa cell expressing mito-Dendra2 from Extended Data Fig. 10m. g-h. Lattice light-sheet time-lapses indicating spread of activated mito-paGFP in control (g) or CytoD-trated (h) cells. i-j. Spinning disk time-lapses of mito-paGFP spread in NT (i) or Arp3 siRNA-treated (j) cells. Time = mm:ss.
Video 10
Simulation of metaphase mitochondrial motility with or without an actin wave. Simulation of damaged (blue) and healthy (red) mitochondrial movement over 30 min in cells with waves (left, black wedge) or without waves (right). Within the wave, 10% of mitochondria undergo comet tail-type motility, while the remaining 90% are immotile to model mitochondria within actin clouds. Outside of the wave, mitochondria undergo diffusion comparable to the motility we observe for mitochondria within the actin cable meshwork. See methods for further details.
Source data
Rights and permissions
About this article
Cite this article
Moore, A.S., Coscia, S.M., Simpson, C.L. et al. Actin cables and comet tails organize mitochondrial networks in mitosis. Nature 591, 659–664 (2021). https://doi.org/10.1038/s41586-021-03309-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03309-5
This article is cited by
-
Exploring the potential link between MitoEVs and the immune microenvironment of periodontitis based on machine learning and bioinformatics methods
BMC Oral Health (2024)
-
Dissecting organelle interdependence
Nature Cell Biology (2024)
-
ClpP/ClpX deficiency impairs mitochondrial functions and mTORC1 signaling during spermatogenesis
Communications Biology (2023)
-
The multiple links between actin and mitochondria
Nature Reviews Molecular Cell Biology (2023)
-
Rationalized deep learning super-resolution microscopy for sustained live imaging of rapid subcellular processes
Nature Biotechnology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.