Abstract
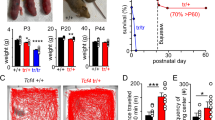
Among numerous challenges encountered at the beginning of extrauterine life, the most celebrated is the first breath that initiates a life-sustaining motor activity1. The neural systems that regulate breathing are fragile early in development, and it is not clear how they adjust to support breathing at birth. Here we identify a neuropeptide system that becomes activated immediately after birth and supports breathing. Mice that lack PACAP selectively in neurons of the retrotrapezoid nucleus (RTN) displayed increased apnoeas and blunted CO2-stimulated breathing; re-expression of PACAP in RTN neurons corrected these breathing deficits. Deletion of the PACAP receptor PAC1 from the pre-Bötzinger complex—an RTN target region responsible for generating the respiratory rhythm—phenocopied the breathing deficits observed after RTN deletion of PACAP, and suppressed PACAP-evoked respiratory stimulation in the pre-Bötzinger complex. Notably, a postnatal burst of PACAP expression occurred in RTN neurons precisely at the time of birth, coinciding with exposure to the external environment. Neonatal mice with deletion of PACAP in RTN neurons displayed increased apnoeas that were further exacerbated by changes in ambient temperature. Our findings demonstrate that well-timed PACAP expression by RTN neurons provides an important supplementary respiratory drive immediately after birth and reveal key molecular components of a peptidergic neural circuit that supports breathing at a particularly vulnerable period in life.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Information or from the corresponding author. Unique biological materials are available from the relevant authors upon reasonable requests. Source data are provided with this paper.
References
Del Negro, C. A., Funk, G. D. & Feldman, J. L. Breathing matters. Nat. Rev. Neurosci. 19, 351–367 (2018).
Guyenet, P. G. et al. The retrotrapezoid nucleus: central chemoreceptor and regulator of breathing automaticity. Trends Neurosci. 42, 807–824 (2019).
Ruffault, P. L. et al. The retrotrapezoid nucleus neurons expressing Atoh1 and Phox2b are essential for the respiratory response to CO2. eLife 4, e07051 (2015).
Holloway, B. B., Viar, K. E., Stornetta, R. L. & Guyenet, P. G. The retrotrapezoid nucleus stimulates breathing by releasing glutamate in adult conscious mice. Eur. J. Neurosci. 42, 2271–2282 (2015).
Weese-Mayer, D. E. et al. An official ATS clinical policy statement: congenital central hypoventilation syndrome: genetic basis, diagnosis, and management. Am. J. Respir. Crit. Care Med. 181, 626–644 (2010).
Shi, Y. et al. Neuromedin B expression defines the mouse retrotrapezoid nucleus. J. Neurosci. 37, 11744–11757 (2017).
Cummings, K. J. et al. Sudden infant death syndrome (SIDS) in African Americans: polymorphisms in the gene encoding the stress peptide pituitary adenylate cyclase-activating polypeptide (PACAP). Acta Paediatr. 98, 482–489 (2009).
Cummings, K. J., Pendlebury, J. D., Sherwood, N. M. & Wilson, R. J. Sudden neonatal death in PACAP-deficient mice is associated with reduced respiratory chemoresponse and susceptibility to apnoea. J. Physiol. (Lond.) 555, 15–26 (2004).
Gray, S. L., Cummings, K. J., Jirik, F. R. & Sherwood, N. M. Targeted disruption of the pituitary adenylate cyclase-activating polypeptide gene results in early postnatal death associated with dysfunction of lipid and carbohydrate metabolism. Mol. Endocrinol. 15, 1739–1747 (2001).
Ross, R. A. et al. PACAP neurons in the ventral premammillary nucleus regulate reproductive function in the female mouse. eLife 7, e35960 (2018).
Rossi, J. et al. Melanocortin-4 receptors expressed by cholinergic neurons regulate energy balance and glucose homeostasis. Cell Metab. 13, 195–204 (2011).
Shi, Y. et al. Nalcn is a “leak” sodium channel that regulates excitability of brainstem chemosensory neurons and breathing. J. Neurosci. 36, 8174–8187 (2016).
Hwang, D. Y., Carlezon, W. A. Jr, Isacson, O. & Kim, K. S. A high-efficiency synthetic promoter that drives transgene expression selectively in noradrenergic neurons. Hum. Gene Ther. 12, 1731–1740 (2001).
Kumar, N. N. et al. Regulation of breathing by CO2 requires the proton-activated receptor GPR4 in retrotrapezoid nucleus neurons. Science 348, 1255–1260 (2015).
Stornetta, R. L. et al. A group of glutamatergic interneurons expressing high levels of both neurokinin-1 receptors and somatostatin identifies the region of the pre-Bötzinger complex. J. Comp. Neurol. 455, 499–512 (2003).
Baertsch, N. A., Baertsch, H. C. & Ramirez, J. M. The interdependence of excitation and inhibition for the control of dynamic breathing rhythms. Nat. Commun. 9, 843 (2018).
Harmar, A. J. et al. Pharmacology and functions of receptors for vasoactive intestinal peptide and pituitary adenylate cyclase-activating polypeptide: IUPHAR review 1. Br. J. Pharmacol. 166, 4–17 (2012).
Arata, S., Nakamachi, T., Onimaru, H., Hashimoto, H. & Shioda, S. Impaired response to hypoxia in the respiratory center is a major cause of neonatal death of the PACAP-knockout mouse. Eur. J. Neurosci. 37, 407–416 (2013).
Dudley, D. J., Branch, D. W., Edwin, S. S. & Mitchell, M. D. Induction of preterm birth in mice by RU486. Biol. Reprod. 55, 992–995 (1996).
Mortola, J. P. Influence of temperature on metabolism and breathing during mammalian ontogenesis. Respir. Physiol. Neurobiol. 149, 155–164 (2005).
Cummings, K. J., Willie, C. & Wilson, R. J. Pituitary adenylate cyclase-activating polypeptide maintains neonatal breathing but not metabolism during mild reductions in ambient temperature. Am. J. Physiol. Regul. Integr. Comp. Physiol. 294, R956–R965 (2008).
Sawczenko, A. & Fleming, P. J. Thermal stress, sleeping position, and the sudden infant death syndrome. Sleep 19 (Suppl.), S267–S270 (1996).
Darnall, R. A. The role of CO2 and central chemoreception in the control of breathing in the fetus and the neonate. Respir. Physiol. Neurobiol. 173, 201–212 (2010).
Willinger, M., James, L. S. & Catz, C. Defining the sudden infant death syndrome (SIDS): deliberations of an expert panel convened by the National Institute of Child Health and Human Development. Pediatr. Pathol. 11, 677–684 (1991).
Goldstein, R. D., Kinney, H. C. & Willinger, M. Sudden unexpected death in fetal life through early childhood. Pediatrics 137, e20154661 (2016).
Filiano, J. J. & Kinney, H. C. A perspective on neuropathologic findings in victims of the sudden infant death syndrome: the triple-risk model. Biol. Neonate 65, 194–197 (1994).
Barrett, K. T. et al. Analysis of PAC1 receptor gene variants in Caucasian and African American infants dying of sudden infant death syndrome. Acta Paediatr. 102, e546–e552 (2013).
Huang, J., Waters, K. A. & Machaalani, R. Pituitary adenylate cyclase activating polypeptide (PACAP) and its receptor 1 (PAC1) in the human infant brain and changes in the Sudden Infant Death Syndrome (SIDS). Neurobiol. Dis. 103, 70–77 (2017).
Paxinos, G. & Franklin, K. B. J. The Mouse Brain in Stereotaxic Coordinates 2nd edn (Elsevier Academic Press, 2004).
Lazarenko, R. M. et al. Acid sensitivity and ultrastructure of the retrotrapezoid nucleus in Phox2b-EGFP transgenic mice. J. Comp. Neurol. 517, 69–86 (2009).
Ting, J. T., Daigle, T. L., Chen, Q. & Feng, G. Acute brain slice methods for adult and aging animals: application of targeted patch clamp analysis and optogenetics. Methods Mol. Biol. 1183, 221–242 (2014).
Liss, B. Improved quantitative real-time RT-PCR for expression profiling of individual cells. Nucleic Acids Res. 30, e89 (2002).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 (2001).
Wiznerowicz, M. & Trono, D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J. Virol. 77, 8957–8961 (2003).
Mortola, J. P. Breathing pattern in newborns. J. Appl. Physiol. 56, 1533–1540 (1984).
Ramirez, S. C. et al. Perinatal breathing patterns and survival in mice born prematurely and at term. Front. Physiol. 10, 1113 (2019).
Pepper, D. R., Landauer, R. C. & Kumar, P. Postnatal development of CO2-O2 interaction in the rat carotid body in vitro. J. Physiol. (Lond.) 485, 531–541 (1995).
Basting, T. M. et al. Hypoxia silences retrotrapezoid nucleus respiratory chemoreceptors via alkalosis. J. Neurosci. 35, 527–543 (2015).
Depuy, S. D., Kanbar, R., Coates, M. B., Stornetta, R. L. & Guyenet, P. G. Control of breathing by raphe obscurus serotonergic neurons in mice. J. Neurosci. 31, 1981–1990 (2011).
Smith, J. C., Ellenberger, H. H., Ballanyi, K., Richter, D. W. & Feldman, J. L. Pre-Bötzinger complex: a brainstem region that may generate respiratory rhythm in mammals. Science 254, 726–729 (1991).
Funk, G. D., Smith, J. C. & Feldman, J. L. Generation and transmission of respiratory oscillations in medullary slices: role of excitatory amino acids. J. Neurophysiol. 70, 1497–1515 (1993).
Ruangkittisakul, A. et al. High sensitivity to neuromodulator-activated signaling pathways at physiological [K+] of confocally imaged respiratory center neurons in on-line-calibrated newborn rat brainstem slices. J. Neurosci. 26, 11870–11880 (2006).
Lorier, A. R. et al. P2Y1 receptor modulation of the pre-Bötzinger complex inspiratory rhythm generating network in vitro. J. Neurosci. 27, 993–1005 (2007).
Acknowledgements
This work was supported by National Institutes of Health (NIH) research grants: R01 HL108609 (D.A.B.), R01 HL074011 (P.G.G.), R01 DK096010 and R01 DK075632 (B.B.L.), and K08 DK118201 (R.A.R.); by grants from CCHS Family Foundation (D.A.B. and P.G.G.), CIHR (159551, G.D.F.), CFI and Stollery Children’s Hospital Foundation through WCHRI (G.D.F. and R.J.R.); and by T32 HL007374 (R.A.R.) and a Harrison Undergraduate Research Award (A.S.). We thank C. B. Bayliss, Y.-H. Chiu, E. Gonye, A. Narahari (Bayliss laboratory) for technical support and suggestions; S. Abbott and G. Souza for suggestions regarding dEMG and plethysmography analysis; J. Hannibal for antibody used to evaluate PACAP re-expression in HEK293T cells; and K. S. Ravichandran for helpful edits of the manuscript.
Author information
Authors and Affiliations
Contributions
Y.T.S., G.D.F., P.G.G. and D.A.B. developed concepts and designed experiments. Y.T.S. developed shRNA constructs, produced lentiviruses, and contributed to all experiments (made viral injections, performed histochemistry, single-cell molecular biology, plethysmography, RTN neuron electrophysiology dEMG recordings and in vivo PACAP injections). D.S.S. performed histochemistry and data analysis. R.J.R. and Y.Z. performed in vitro rhythmic slice recordings, which were also analysed by G.D.F. A.S., Y.W., A.N. and K.Y.L. analysed histochemical and plethysmography data; K.Y.L. and Y.T.S. made adult mouse brainstem slices. E.P.R. designed viral targeting constructs and subcloned shRNA. R.A.R. and B.B.L. provided mice with floxed Pacap alleles. R.L.S. analysed and interpreted histochemical data. Y.T.S. and D.A.B. conceptualized and wrote the manuscript, and all authors edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Gilles Fortin, Jan Ramirez and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Validation of PACAP recombination in RTN neurons from Phox2b::cre;Pacapfl/fl mice.
a, Multiplex in situ hybridization for Pacap and Nmb in the ventral brainstem of Phox2b::cre;Pacapfl/fl mice that were either Cre-negative (left) or Cre-positive (right). The boxed areas in low-power fluorescence images (middle) are expanded in higher power images (bottom) to reveal selective Pacap deletion from Nmb-expressing RTN neurons in Cre-positive mice, with no effect on Pacap expression in midline regions. Nmb+ neurons indicated with an asterisk are expanded in insets; arrow indicates non-Nmb+ cell that expresses Pacap. Data representative of two replicate experiments from each of three Cre-negative and Cre-positive mice. Coronal section; scale bars, 200 μm and 100 μm. b, Single-cell RT–PCR of GFP-labelled RTN neurons, verified by expression of Phox2b and Nmb, from control Cre-negative Pacapfl/fl mice (lanes 1–3) and Phox2b::cre;Pacapfl/fl mice (lanes 4–6). Pacap transcripts from Cre-positive RTN neurons were smaller, as expected for excision of exon 2; when recombined, this excision creates a frameshift mutation10. Data representative of 16 cells from Cre-negative and Cre-positive mice (n = 2 each). c, d, Effects of raised ambient CO2 (FiCO2, 4–8%; balance O2) on respiratory frequency (c; mean ± s.e.m.) and tidal volume (d; VT, mean ± s.e.m.) in Pacapfl/fl (n = 23) and Phox2b::cre;Pacapfl/fl mice (n = 12). VT: F1,33 = 0.5134, P = 0.4787; fR: F1,33 = 10.22, P = 0.0031, **P = 0.0013, ***P = 0.0005 for Pacapfl/fl versus cre;Pacapfl/fl by two-way repeated measures ANOVA, Sidak multiple comparison. e, Ventilation in varied ambient O2 (normoxia: FiO2, 21%; hypoxia: FiO2, 10%; hyperoxia: FiO2, 100%) for Pacapfl/fl (n = 23) and Phox2b::cre;Pacapfl/fl mice (n = 12). F1,99 = 5.332, P = 0.0230, for Pacapfl/fl versus cre;Pacapfl/fl by two-way ANOVA, Sidak multiple comparison. Box plots as in Fig. 1c.
Extended Data Fig. 2 Effects of Cre-mediated deletion of PACAP in RTN neurons of adult mice on breathing and CO2-evoked neuronal activation in vivo.
a, Schematic depicting approach for PRSx8 promoter-driven lentiviral injections in Pacapfl/fl mice using stereotaxic coordinates and antidromic facial field potentials to locate the RTN. Outline of experimental design for recording physiological responses before and 4 weeks after lentivirus injections. b, Virally-infected (mCherry+) RTN neurons (Nmb+) in Phox2b::cre;Pacapfl/fl mice injected with control or PACAP-expressing lentivirus. Coronal section; scale bar, 100 μm (n = 9 mice each). c, Quantification of viral transduction specificity (percentage of mCherry-labelled neurons in the RTN region that were Nmb+) and efficiency (percentage of Nmb-expressing neurons that were mCherry+) in mice injected with lentivirus expressing mCherry (control) or PACAP (n = 9 each), or mCherry (no Cre) and Cre-mCherry (n = 5 each). d, RNAscope for Pacap, Nmb and Th in the caudal RTN region of the rostral ventrolateral medulla (−6.5 mm, relative to bregma); boxed area from low-power merged image is expanded (right) to show that Nmb-expressing RTN neurons universally express PACAP (yellow arrows) whereas only few Th-expressing C1 neurons express Pacap in this region and most Pacap-expressing C1 cells are located mediodorsal to the RTN (purple arrows). Pacap is also expressed in non-RTN, non-C1 cells (red arrowheads). Representative of three independent experiments (as quantified in e, f). Coronal section; scale bars, 100 μm. e, f, Cell counts from in situ hybridization experiments (n = 3) through the C1 region quantifying Th-expressing C1 neurons that also express Pacap (e; mean ± s.e.m.), and the percentage of Pacap-expressing C1 neurons (f; mean ± s.e.m.). Note that most neurons in the caudal C1 express Pacap, whereas progressively fewer rostral C1 neurons express Pacap in the region that overlaps with the RTN. (‘X ‘marks the approximate rostrocaudal level depicted in d.) g, RNAscope for Pacap and Nmb combined with immunostaining for mCherry in the RTN region of Pacapfl/fl mice that were injected with virus for mCherry or Cre-mCherry (schematic on right). Transduced RTN neurons are indicated by yellow arrows and uninfected RTN neurons by green arrows. Coronal section; scale bar, 100 μm. h. Effects of raised ambient CO2 (FiCO2, 4–8%; balance O2) on ventilation (mean ± s.e.m.), respiratory frequency (mean ± s.e.m.) and tidal volume (mean ± s.e.m.) in Pacapfl/fl injected in the RTN with control (mCherry) or Cre-mCherry lentivirus. Phox2b::cre;Pacapfl/fl mice (n = 5 each). The 95% confidence intervals are from all mice before virus injection. VE: F2,68 = 47.38, VT: F2,68 = 16.22, fR: F2,68 = 23.05, all P < 0.0001 for treatment main effects; *P < 0.05, **P < 0.005, ***P < 0.001, ****P < 0.0001 for Cre versus initial; +P < 0.05, ++P < 0.005, ++++P < 0.0001 for Cre versus no Cre, by two-way ANOVA, Tukey’s multiple comparison. i, j, CO2-evoked change in VE (i) and apnoea frequency (j) in Pacapfl/fl mice before and 4 weeks after RTN injection of control (mCherry) or Cre-mCherry lentivirus (n = 5 each; as in Fig. 1d). ∆VE: F3,16 = 11.39, P = 0.0003: apnoea: F3,16 = 34.3, P < 0.0001 by ANOVA; **P = 0.012 (i) and ****P < 0.0001 (j) Cre-mCherry versus control or pre-injection, Tukey’s multiple comparison. Box plots as in Fig. 1c. The shaded regions depict 95% confidence intervals for uninjected Pacapfl/fl mice. k, Combined RNAscope for Nmb and Fos, and immunostaining for mCherry, Pacapfl/fl mice injected with lentivirus for control (mCherry) or Cre-mCherry after exposure to CO2 (12% CO2, 60% O2, balance N2 for 35 min). CO2-induced Fos expression was observed in numerous uninfected (green arrows) and virally transduced RTN neurons (yellow arrows). For both control and Cre-mCherry (n = 5 each). The percentage of CO2-activated RTN neurons (mean ± s.e.m.) was not different in uninfected and infected RTN neurons from control- or Cre-mCherry-injected mice. Coronal section; scale bar, 100 μm.
Extended Data Fig. 3 Breathing is suppressed by PACAP knockdown in RTN neurons.
a, Top, lentiviral construct incorporating PACAP (or control) shRNA sequence at a Drosha site derived from miR30a was embedded in an artificial intron upstream of an mCherry coding sequence and driven by a PRSx8 promoter. Bottom, schematic illustrating lentiviral injection approach and follow up validation of knockdown and respiratory assessment. b, Quantification of viral transduction specificity (percentage of mCherry-labelled neurons in the RTN region that were GFP+) and efficiency (percentage of GFP-labelled neurons that were mCherry+) in a subset of mice expressing control (n = 6) or PACAP shRNA (n = 10). Box plots as in Fig. 1c. c, d, In situ hybridization combined with GFP and mCherry immunohistochemistry showing expression of transcripts for Pacap (c) or Vglut2 (d) in RTN neurons transduced with lentivirus expressing control shRNA (top) or Pacap shRNA (bottom). Transduced RTN neurons indicated by yellow arrows, uninfected RTN neurons by green arrows. Data representative of two replicate experiments from mice injected with virus for control (n = 6) and Pacap shRNA (n = 10). Coronal section; scale bars, 100 μm. e, Schematic shows single GFP-expressing RTN neurons aspirated under direct vision into pipettes from brainstem slices, and processed for multiplex qRT–PCR (top). Cumulative frequency distribution of Pacap (middle) and Vglut2 (bottom) transcript levels from uninjected mice (GFP), or mice infected with either control or Pacap shRNA. Red arrow indicates percentage of cells infected with Pacap shRNA in which Pacap expression was undetectable. Insets, individual cell expression levels for Pacap and Vglut2 (relative to Gapdh; median and 95% confidence intervals, with number of cells: number of mice). ****P < 0.0001 versus GFP and control (CON), by Kruskal–Wallis one-way ANOVA. f, Effects of raised ambient CO2 (FiCO2, 4–8%; balance O2) on respiratory frequency and tidal volume (mean ± s.e.m.) from mice before and 4 weeks after RTN injection with either control (n = 9) or Pacap (n = 13) shRNA-expressing lentivirus. fR: F3,160 = 6.68, P < 0.0001, for treatment by two-way ANOVA; *P = 0.0414 in 4% and P = 0.0272 in 8% CO2, **P = 0.0011, for PACAP shRNA versus initial (Tukey’s multiple comparison). VT: F3,160 = 9.94, P < 0.0001, for treatment by two-way ANOVA. *P = 0.0115, **P = 0.0011, for Pacap shRNA versus initial (Tukey’s multiple comparison). g, CO2-evoked change in ventilation in Phox2b::GFP mice before and 4 weeks after RTN injection of control (n = 9) or Pacap shRNA (n = 13) expressing lentivirus. ***P = 0.0002 by two-sided Wilcoxon matched-pairs signed-rank test. h, Ventilation in varied ambient O2 (normoxia: FiO2, 21%; hypoxia: FiO2, 10%; hyperoxia: FiO2, 100%) from mice 4 weeks after RTN injection with either control (n = 9) or PACAP (n = 13) shRNA-expressing lentivirus. F3,120 = 0.1761, P = 0.9124, for treatment (control versus Pacap shRNA) by two-way ANOVA.
Extended Data Fig. 4 CO2 sensitivity of RTN and preBötC neurons after PACAP depletion.
a, Firing rate histogram from cell-attached recording of RTN neuron exposed to acidified and alkalized bath solutions in brainstem slice from a Cre-positive Phox2b::cre;Pacapfl/fl mouse. b, The pH sensitivity of RTN neurons (pH50, pH where firing rate is half that obtained at pH 7.0) obtained from Phox2b::cre;Pacapfl/fl mice that were Cre-negative or Cre-positive. (n cells: N mice; P = 0.8248 by unpaired t-test). Box plots as in Fig. 1c. c, Single-cell qRT–PCR for the indicated transcripts from RTN neurons collected after recordings of pH sensitivity in brainstem slices from Cre− or Cre+ Phox2b::cre;Pacapfl/fl mice (n cells: N mice); F3,80 = 7.729, P = 0.0001 for interaction by two-way ANOVA, ****P < 0.0001 for PACAP from Cre− versus Cre+ mice. d, Left, mice injected with lentivirus for control (top) or Pacap shRNA (bottom) were exposed to CO2 (12% CO2, 60% O2, balance N2 for 45 min). Immunohistochemistry for GFP and mCherry (left), and Fos (right), reveals CO2-induced Fos expression in numerous uninfected (green arrows) and lentiviral-transduced RTN neurons (yellow arrows). Right, percentage of Fos-immunopositive expressing RTN neurons (mean ± s.e.m.) under control conditions (0% CO2) and after CO2 exposure in uninjected mice (n = 3 each), and in mice injected with lentivirus expressing either control or PACAP shRNA (n = 3 each). Coronal section; scale bars, 100 μm. e, GFP immunohistochemistry for PHOX2B-expressing RTN neurons combined with in situ hybridization for Pac1 receptor. Filled arrow, PHOX2B-expressing RTN neuron associated with Pac1; filled arrowheads, PHOX2B-expressing RTN neurons without Pac1; empty arrow, Pac1-expressing neuron outside the RTN. Data representative of experiments from three mice. Coronal section; scale bar, 100 μm. f, Top, Fos immunolabelling in coronal brainstem sections from uninjected mice under control conditions and after CO2 exposure, and in CO2-exposed mice that had received RTN injections of lentivirus expressing scrambled or Pacap shRNA. Scale bar, 100 μm. Bottom, quantification of Fos-immunolabelled cells in the preBötC region. Mean cell counts (± s.e.m.) were obtained from several sections in either coronal or parasagittal plane from individual mice and treated as a single data point for subsequent analysis (n = mice analysed for each condition). Data from coronal and parasagittal sections were essentially identical and combined for subsequent analysis. For coronal: F3,8 = 22.4, P = 0.003 by ANOVA; control versus CO2, P = 0.0007; control versus CO2+sc, P = 0.0027; CO2+sh vs CO2, P = 0.0015; CO2+sh vs CO2+sc, P = 0.0060, Tukey’s multiple comparison. For parasagittal: F3,10 = 27.25, P < 0.0001 by ANOVA; control versus CO2, P < 0.0001; control versus CO2+sc, P = 0.0003; CO2+sh versus CO2, P = 0.0009; CO2+sh vs CO2+sc, P = 0.0030, Tukey’s multiple comparison. ‘sc’ denotes scrambled shRNA; ‘sh’ denotes Pacap shRNA. g, Schematic in the parasagittal plane illustrating the location of CO2/H+-sensitive chemoreceptor neurons in the RTN and their projections to respiratory regions of the brainstem. KF, Kölliker-Fuse; LBP, lateral parabrachial nucleus; NTS, nucleus tractus solitarius. h, Immunofluorescence labelling for mCherry and ChAT combined with multiplex in situ hybridization for Pac1, Sst and/or Fos in coronal sections containing the preBötC from mice injected in the RTN with lentivirus expressing control or Pacap shRNA. The preBötC was identified by its location relative to the nucleus ambiguus (that is, see ChAT-IR) in a region with a high concentration of Sst-expressing neurons. The preBötC is innervated by fibres from lentivirus-infected RTN neurons (see mCherry staining), and contains cells that express PAC1. After exposure to elevated CO2 (12%, 60% O2, balance N2 for 35 min) preBötC cells express Fos. The data are representative of two replicate experiments from each of 6 mice tested for CO2-induced Fos expression after control lentivirus injection in RTN. Scale bar, 100 μm. i, Histochemical localization of PAC1 and CO2-induced Fos expression together with markers for neurons (Map2) or astrocytes (Aldh1l1), and for excitatory (Vglut2) or inhibitory neurons (Gad1, Gad2). Insets, merged images from boxed regions at higher magnification. Filled arrows, Pac1- and Fos-expressing cells containing the marker; open arrows: Pac1- and Fos-expressing cells without the marker; filled arrowhead: Fos with no Pac1 or marker; empty arrowhead: Pac1 cells containing the marker but no Fos. Data representative of two replicate experiments from each of four mice. Coronal section; scale bar, 100 μm.
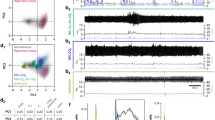
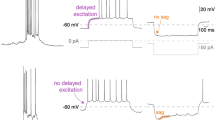
Extended Data Fig. 5 Effect of PACAP injection in the preBötC on respiratory output.
a, Summary data illustrating effects of vehicle (PBS) and PACAP injections in the preBötC (n = 15 injections, n = 6 anaesthetized mice) on dEMG burst frequency and burst amplitude. Data were normalized to control values before PBS. Box plots as in Fig. 1c. Repeated-measures ANOVA on absolute values for frequency; F2,28 = 62.71, ****P < 0.0001 for PACAP versus control and PBS; and for amplitude: F2,28 = 13.39, ***P = 0.0002 and P = 0.0005 for PACAP versus control and PBS, Tukey’s multiple comparison. b, Injection locations mapped onto ventral brainstem cutouts from images of parasagittal sections at two levels (1.2 and 1.32 mm lateral, from figures 111 and 112 of ref. 29), colour coded for effect size. The approximate location of the preBötC (orange shaded) is based on the area caudal to facial motor nucleus, rostral to lateral reticular nucleus; and ventral to nucleus ambiguus containing neuronal SST expression (not shown). c, Rate histogram of inspiratory burst frequency and integrated dEMG during vehicle, DLH (40 pmol) and PACAP (0.3 pmol) injection into the ipsilateral and contralateral preBötC of an anaesthetized mouse 4 weeks after on-target unilateral injection of Pac1 shRNA-expressing lentivirus (ipsilateral data reproduced from Fig. 3b, ipsilateral injection sites shown in Fig. 3d). d, Top, photomicrograph from parasagittal brainstem section of the single mouse (of n = 6) with an off-target injection of Pac1 shRNA lentivirus (that is, outside the preBötC). The approximate boundary of virally-transduced cells (mCherry-expressing) outside the preBötC (red dashed area) and PACAP injection site in the preBötC (green) were mapped onto ventral brainstem cutout (from figure 111 of ref. 29). Parasagittal section; scale bar, 200 μm. Bottom, corresponding rate histogram of inspiratory burst frequency and integrated dEMG during vehicle, DLH (40 pmol) and PACAP (0.3 pmol) injection into the ipsilateral preBötC. e, Time course depicting the effect on hypoglossal nerve burst frequency (mean ± s.e.m.) of pressure injections of PACAP (1 μM, 10 μM) into the preBötC of rhythmic brain slice preparations prepared from two age groups of neonatal mice (P0–P3, n = 8 slices; P9–P10, n = 7 slices). f, Peak relative hypoglossal nerve burst frequency after PACAP application (1 uM and 10 μM) in rhythmic brain slice preparations were not different in two age groups of neonatal mice (P0–P3, P9–P10); data from all ages were pooled for statistical analysis. *P = 0.0147, ****P < 0.0001 relative to baseline, by ANOVA. g, Rhythmic brain slice (P0) was bathed in 5 mM K+ to reduce network excitability and the effect of pressure injection of PACAP (100 nM) on hypoglossal nerve burst activity was determined. Inset, peak relative hypoglossal nerve burst frequency after 100 nM PACAP injection in slices bathed in 5 mM K+ (P0–P3, n = 6). h. Multiplex in situ hybridization for Sst and Pac1 transcripts combined with ChAT immunostaining in parasagittal brainstem sections containing the preBötC from mice at the indicated embryonic and postnatal ages. Images representative of (mice:litters): E19.5 (12:3), P1 (11:3), P4 (8:3), P7 (8:3), P14 (8:3), and P120 (3). Parasagittal sections; scale bar, 100 μm. i, RNAscope for Sst and either Vpac1 (also known as V1pr1; top) or Vpac2 (V1pr2; bottom) transcripts combined with ChAT immunostaining in parasagittal brainstem sections containing the preBötC from mice at the indicated embryonic and postnatal ages. Note the absence of either VPAC1 or VPAC2 expression in preBötC neurons, with a developmental increase in VPAC2 transcripts evident in ChAT-IR neurons of the nucleus ambiguus beginning around P7 and persisting into adulthood (arrows). Images representative of (mice:litters): E19.5 (9:3), P1 (9:3), P4 (6:3), P7 (6:3), P14 (6:3), and P120 (3) Parasagittal sections; scale bar, 100 μm.
Extended Data Fig. 6 Localization of Pac1 shRNA lentiviral transduction sites in the preBötC and effects on CO2-stimulated breathing.
a, b, Top, bilateral fluorescent images of transduction region overlaid on dark field images of parasagittal brainstem sections from mice injected with PAC1 shRNA-expressing (a) and control (b) lentivirus. Parasagittal sections, scale bars, 500 μm. Lower, left: viral transduction regions from the two images were superimposed on a schematic of the relevant brainstem region (adapted from plate 111 of ref. 29). The preBötC region (orange shaded) was approximated based on a similar tracing of concentrated Sst expression (not shown), and by its relationship to select anatomical features. Bottom right, the intersection of the viral transduction region with the preBötC region was determined from each of these tracings (and from a second set of bilateral images, corresponding to plate 112 of ref. 29), and averaged to obtain a percentage overlap with the preBötC. The effect of increasing concentrations of CO2 (balance O2) on ventilation was measured before and 4 weeks after preBötC injection of PAC1 shRNA-expressing lentivirus, and the percentage reduction in CO2-stimulated breathing determined (at 8% CO2). c, Percentage overlap of the viral transduction region with the preBötC in mice injected with control or Pac1 shRNA-expressing lentivirus (n = 7 and 10). P = 0.07, by Mann–Whitney t-test. Box plots as in Fig. 1c. d, Effects of raised ambient CO2 (FiCO2, 4% to 8%; balance O2) on respiratory frequency (and tidal volume (mean ± s.e.m.) before and 4 weeks after preBötC injection with either control (n = 9) or Pac1 (n = 11) shRNA-expressing lentivirus. VT: F3,144 = 0.612, P = 0.6082; fR: F3,144 = 7.761, P < 0.0001, for condition by two-way ANOVA. *P < 0.05, PAC1 shRNA versus initial; ++P < 0.01, Pac1 shRNA versus control shRNA, Tukey’s multiple comparison. e, CO2-evoked change in ventilation in Phox2b::GFP mice before and 4 weeks after preBötC injection of control (n = 9) or Pac1 shRNA (n = 13) expressing lentivirus. **P = 0.0039, two-sided Wilcoxon matched-pairs signed-rank test. f, Ventilation in varied ambient O2 (normoxia: FiO2, 21%; hypoxia: FiO2, 10%; hyperoxia: FiO2, 100%; median, 25–75th percentile with minima and maxima) from mice before and 4 weeks after RTN injection with either control (n = 9) or Pacap (n = 13) shRNA-expressing lentivirus. F3,108 = 1.204, P = 0.3120, for treatment by two-way ANOVA.
Extended Data Fig. 7 Developmental expression of PACAP and GAPDH in RTN neurons.
a, Expression of Pacap, Nmb and Vglut2 in RTN neurons assessed by multiplex in situ hybridization from RTN neurons at the indicated postnatal ages. Images representative of (mice:litters): E17.5 (17:3), E19.5 (18:3), P0 (12:3), P3 (10:3), P6 (9:3), P14 (9:3), P50 (5), and P120 (4). Coronal sections; scale bar, 100 μm. b, Left, cumulative probability distribution for individual Pacap expression values (\({2}^{-\Delta {C}_{{\rm{t}}}}\), relative to Gapdh) from single-cell qRT–PCR of RTN neurons at four different embryonic and postnatal ages (cells:mice): E17.5–E19.5 (97:20); P0-P3 (150:20); P4–P14 (218:24); P52–P200 (114:14). Inset, individual cell Pacap expression levels within each group, relative to Gapdh (\({2}^{-\Delta {C}_{{\rm{t}}}}\)). Box plots as in Fig. 1c. ****P < 0.0001 versus E17.5–E19.5, ++++P < 0.0001 versus P0–P3, Kruskal–Wallis one-way ANOVA with Dunn’s multiple comparisons test. Right, grouped box plot of median values for each of the embryonic and postnatal ages (from raw data presented in Fig. 4b). c, Expression of Phox2b, Nmb and Vglut2 assessed by multiplex sc-qPCR from RTN neurons collected from brainstem slices at the indicated postnatal ages (cells:mice): E17.5 (14:6); P1 (16:3); P3 (18:3); P5 (17:2); P8 (14:2); P11 (29:3); P50 (15:2); P100 (25:3). *P = 0.0176, **P = 0.0026, by Kruskal–Wallis ANOVA with multiple Dunn’s post hoc comparisons to P1. d, RNAscope indicated postnatal ages for Pacap expression in Th-expressing neurons in the caudal C1 region (−6.9 mm, relative to bregma) and in Phox2b-expressing NTS neurons (−7.1 mm, relative to bregma). Images representative of (mice:litters): E18.5 (12:3), E19.5 (12:3), P0 (10:3), P1 (8:3), and P5 (8:3). Coronal sections, scale bars, 100 μm.
Extended Data Fig. 8 Pacap expression in Nmb-expressing RTN neurons from pre-term mice.
a, Multiplex in situ hybridization for Pacap and Nmb in RTN neurons from pre-term mice born at E17.5, E18.5 and E19.5. Newborn pups were extracted from the amniotic sac by the dam (top row), retained in the amniotic sac until transcardial perfusion (middle row), or obtained after one day of postnatal life (bottom row, P1). The data are representative of (mice:litters): E17.5, P0o (12:3), P0i (6:3), P1 (6:3); E18.5, P0o (12:3), P0i (5:3), P1 (6:3); E19.5, P0o (13:3), P0i (4:3), P1 (6:3). Coronal sections; scale bar, 100 μm. b, Expression of Phox2b, Nmb and Vglut2 assessed by multiplex sc-qPCR from RTN neurons obtained from brainstem slices of preterm mouse pups (E17.5–E19.5) at the indicated postnatal days (P0–P2); at birth (P0), some pups were taken en caul (in the amniotic sac; P0i) or after maternal extraction from the amniotic sac (P0o) (n:N values as in Fig. 4e). Box plots as in Fig. 1c.
Extended Data Fig. 9 CO2-stimulated breathing and apnoea incidence in conditional PACAP-knockout mice across the early postnatal period and during thermal stress.
a, Ventilation in hyperoxia (60% O2, balance N2) and in hyperoxic hypercapnia (10% CO2, 60% O2, balance N2) in Phox2b::cre;Pacapfl/fl mice and Cre-negative, Pacapfl/fl control littermates (n = 14 pups each, from 3 litters) at different early postnatal ages (P2–P5, P6–P9, P10–P12). ****P < 0.0001, for genotype by two-way repeated-measures ANOVA, Sidak’s multiple comparison. b, Summary data of the CO2-induced change in ventilation at the indicated early postnatal ages for Phox2b::cre;Pacapfl/fl mice and Cre-negative, Pacapfl/fl control littermates (P2–P5, n = 14 each; P6–P9, n = 14 each; P10–P12, n = 11 and 13). Box plots as in Fig. 1c. F1,74 = 78.42, P < 0.0001, for genotype by two-way ANOVA. ****P < 0.0001 genotype effect for age group, Sidak’s multiple comparison. c, Summary data of apnoea incidence in Phox2b::cre;Pacapfl/fl mice and Cre-negative, Pacapfl/fl control littermates at different early postnatal ages (P2–P5, n = 13 and 14; P6–P9, n = 14 each; P10–P12, n = 13 each). F1,75 = 69.13, P < 0.0001, for genotype by two-way ANOVA. ***P = 0.0002, ****P < 0.0001 genotype effect for age group, Sidak’s multiple comparison. d, Summary data for the CO2-induced change in ventilation, tidal volume and frequency in Phox2b::cre;Pacapfl/fl mice and Cre-negative, Pacapfl/fl control littermates (n = 14 pups each, from 3 litters); data from individual mice at different ages (P2–P5, P6–P9, P10–P12) were averaged, and treated as a single data point for analysis between genotypes. ****P < 0.0001, ***P = 0.0002, **P = 0.0032, unpaired t-test. e, Summary data of apnoea incidence in Phox2b::cre;Pacapfl/fl mice and Cre-negative, Pacapfl/fl control littermates at different ages (P2–P5, P6–P9, P10–P12); mice were studied in normoxia at the indicated ambient temperatures (N, number of mice for each age and condition). ****P < 0.0001, for genotype effect by two-way ANOVA; +P = 0.0143, ++P = 0.0059, +++P = 0.0002, ++++P < 0.0001, comparing Cre− versus Cre+ at the indicated ambient temperatures, Sidak’s multiple comparison.
Supplementary information
Supplementary Tables
This file provides primer and probe sequences used for RT-PCR, shRNA and RNAscope in situ hybridization, and a list of antibodies used for immunohistochemistry.
Source data
Rights and permissions
About this article
Cite this article
Shi, Y., Stornetta, D.S., Reklow, R.J. et al. A brainstem peptide system activated at birth protects postnatal breathing. Nature 589, 426–430 (2021). https://doi.org/10.1038/s41586-020-2991-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2991-4
This article is cited by
-
Galaninergic and hypercapnia-activated neuronal projections to the ventral respiratory column
Brain Structure and Function (2024)
-
Unexplained Significant Central Sleep Apnea in Infants: Clinical Presentation and Outcomes
Sleep and Breathing (2023)
-
Transcriptomes of electrophysiologically recorded Dbx1-derived respiratory neurons of the preBötzinger complex in neonatal mice
Scientific Reports (2022)
-
Distribution of PACAP and PAC1 Receptor in the Human Eye
Journal of Molecular Neuroscience (2022)
-
Gain-of-function mutations in KCNK3 cause a developmental disorder with sleep apnea
Nature Genetics (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.