Abstract
Zygotic genome activation (ZGA) is the first transcription event in life1. However, it is unclear how RNA polymerase is engaged in initiating ZGA in mammals. Here, by developing small-scale Tn5-assisted chromatin cleavage with sequencing (Stacc–seq), we investigated the landscapes of RNA polymerase II (Pol II) binding in mouse embryos. We found that Pol II undergoes ‘loading’, ‘pre-configuration’, and ‘production’ during the transition from minor ZGA to major ZGA. After fertilization, Pol II is preferentially loaded to CG-rich promoters and accessible distal regions in one-cell embryos (loading), in part shaped by the inherited parental epigenome. Pol II then initiates relocation to future gene targets before genome activation (pre-configuration), where it later engages in full transcription elongation upon major ZGA (production). Pol II also maintains low poising at inactive promoters after major ZGA until the blastocyst stage, coinciding with the loss of promoter epigenetic silencing factors. Notably, inhibition of minor ZGA impairs the Pol II pre-configuration and embryonic development, accompanied by aberrant retention of Pol II and ectopic expression of one-cell targets upon major ZGA. Hence, stepwise transition of Pol II occurs when mammalian life begins, and minor ZGA has a key role in the pre-configuration of transcription machinery and chromatin for genome activation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data have been deposited to GEO with the accession number GSE135457.
Code availability
Software and code used to analyse these data are listed in the Nature Research Reporting Summary and are all publicly available.
References
Aoki, F., Worrad, D. M. & Schultz, R. M. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev. Biol. 181, 296–307 (1997).
Roeder, R. G. 50+ years of eukaryotic transcription: an expanding universe of factors and mechanisms. Nat. Struct. Mol. Biol. 26, 783–791 (2019).
Matsui, T., Segall, J., Weil, P. A. & Roeder, R. G. Multiple factors required for accurate initiation of transcription by purified RNA polymerase II. J. Biol. Chem. 255, 11992–11996 (1980).
Zhou, Q., Li, T. & Price, D. H. RNA polymerase II elongation control. Annu. Rev. Biochem. 81, 119–143 (2012).
Nechaev, S. & Adelman, K. Pol II waiting in the starting gates: regulating the transition from transcription initiation into productive elongation. Biochim. Biophys. Acta. 1809, 34–45 (2011).
Core, L. & Adelman, K. Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev. 33, 960–982 (2019).
Kwak, H., Fuda, N. J., Core, L. J. & Lis, J. T. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339, 950–953 (2013).
Adelman, K. & Lis, J. T. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet. 13, 720–731 (2012).
Jukam, D., Shariati, S. A. M. & Skotheim, J. M. Zygotic genome activation in vertebrates. Dev. Cell 42, 316–332 (2017).
Abe, K. et al. The first murine zygotic transcription is promiscuous and uncoupled from splicing and 3′ processing. EMBO J. 34, 1523–1537 (2015).
Kwasnieski, J. C., Orr-Weaver, T. L. & Bartel, D. P. Early genome activation in Drosophila is extensive with an initial tendency for aborted transcripts and retained introns. Genome Res. 29, 1188–1197 (2019).
Blythe, S. A. & Wieschaus, E. F. Zygotic genome activation triggers the DNA replication checkpoint at the midblastula transition. Cell 160, 1169–1181 (2015).
Vastenhouw, N. L. et al. Chromatin signature of embryonic pluripotency is established during genome activation. Nature 464, 922–926 (2010).
Schultz, R. M. The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Hum. Reprod. Update 8, 323–331 (2002).
Abe, K. I. et al. Minor zygotic gene activation is essential for mouse preimplantation development. Proc. Natl Acad. Sci. USA 115, E6780–E6788 (2018).
Kaya-Okur, H. S. et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10, 1930 (2019).
Carter, B. et al. Mapping histone modifications in low cell number and single cells using antibody-guided chromatin tagmentation (ACT-seq). Nat. Commun. 10, 3747 (2019).
Wang, Q.H. et al. CoBATCH for high-throughput single-cell epigenomic profiling. Mol. Cell 76, 206–216 (2019).
Ku, W. L. et al. Single-cell chromatin immunocleavage sequencing (scChIC-seq) to profile histone modification. Nat. Methods 16, 323–325 (2019).
Skene, P. J. & Henikoff, S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6, e21856 (2017).
Abe, K., Inoue, A., Suzuki, M. G. & Aoki, F. Global gene silencing is caused by the dissociation of RNA polymerase II from DNA in mouse oocytes. J. Reprod. Dev. 56, 502–507 (2010).
Zuccotti, M., Piccinelli, A., Giorgi Rossi, P., Garagna, S. & Redi, C. A. Chromatin organization during mouse oocyte growth. Mol. Reprod. Dev. 41, 479–485 (1995).
Tokoro, M. et al. Deposition of acetylated histones by RNAP II promoter clearance may occur at onset of zygotic gene activation in preimplantation mouse embryos. J. Reprod. Dev. 56, 607–615 (2010).
Inoue, A., Jiang, L., Lu, F., Suzuki, T. & Zhang, Y. Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 547, 419–424 (2017).
Wu, J. et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 534, 652–657 (2016).
Wozniak, G. G. & Strahl, B. D. Hitting the ‘mark’: interpreting lysine methylation in the context of active transcription. Biochim. Biophys. Acta. 1839, 1353–1361 (2014).
Gross, D. S. & Garrard, W. T. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 57, 159–197 (1988).
Zheng, H. et al. Resetting epigenetic memory by reprogramming of histone modifications in mammals. Mol. Cell 63, 1066–1079 (2016).
Smith, Z. D. & Meissner, A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 14, 204–220 (2013).
Nguyen, V. T. et al. In vivo degradation of RNA polymerase II largest subunit triggered by α-amanitin. Nucleic Acids Res. 24, 2924–2929 (1996).
Peaston, A. E. et al. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev. Cell 7, 597–606 (2004).
Dubois, M. F., Nguyen, V. T., Bellier, S. & Bensaude, O. Inhibitors of transcription such as 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole and isoquinoline sulfonamide derivatives (H-8 and H-7*) promote dephosphorylation of the carboxyl-terminal domain of RNA polymerase II largest subunit. J. Biol. Chem. 269, 13331–13336 (1994).
Huynh, K. D. & Lee, J. T. Imprinted X inactivation in eutherians: a model of gametic execution and zygotic relaxation. Curr. Opin. Cell Biol. 13, 690–697 (2001).
Jung, Y. H. et al. Maintenance of CTCF- and transcription factor-mediated interactions from the gametes to the early mouse embryo. Mol. Cell 75, 154–171.e5 (2019).
Zhang, B. et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature 537, 553–557 (2016).
Shirane, K. et al. Mouse oocyte methylomes at base resolution reveal genome-wide accumulation of non-CpG methylation and role of DNA methyltransferases. PLoS Genet. 9, e1003439 (2013).
Wang, L. et al. Programming and inheritance of parental DNA methylomes in mammals. Cell 157, 979–991 (2014).
Xu, Q. et al. SETD2 regulates the maternal epigenome, genomic imprinting and embryonic development. Nat. Genet. 51, 844–856 (2019).
Ferguson-Smith, A. C. Genomic imprinting: the emergence of an epigenetic paradigm. Nat. Rev. Genet. 12, 565–575 (2011).
Wang, C. et al. Reprogramming of H3K9me3-dependent heterochromatin during mammalian embryo development. Nat. Cell Biol. 20, 620–631 (2018).
Veselovska, L. et al. Deep sequencing and de novo assembly of the mouse oocyte transcriptome define the contribution of transcription to the DNA methylation landscape. Genome Biol. 16, 209 (2015).
Dahl, J. A. et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 537, 548–552 (2016).
Ge, S. X. Exploratory bioinformatics investigation reveals importance of “junk” DNA in early embryo development. BMC Genomics 18, 200 (2017).
Xu, Q. & Xie, W. Epigenome in early mammalian development: inheritance, reprogramming and establishment. Trends Cell Biol. 28, 237–253 (2018).
Eckersley-Maslin, M. A., Alda-Catalinas, C. & Reik, W. Dynamics of the epigenetic landscape during the maternal-to-zygotic transition. Nat. Rev. Mol. Cell Biol. 19, 436–450 (2018).
Hsu, H. T. et al. Recruitment of RNA polymerase II by the pioneer transcription factor PHA-4. Science 348, 1372–1376 (2015).
Chronis, C. et al. Cooperative binding of transcription factors orchestrates reprogramming. Cell 168, 442–459 (2017).
Yue, F. et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature 515, 355–364 (2014).
Inoue, A., Akiyama, T., Nagata, M. & Aoki, F. The perivitelline space-forming capacity of mouse oocytes is associated with meiotic competence. J. Reprod. Dev. 53, 1043–1052 (2007).
Schmid, M., Durussel, T. & Laemmli, U. K. ChIC and ChEC; genomic mapping of chromatin proteins. Mol. Cell 16, 147–157 (2004).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protocols 9, 171–181 (2014).
Brind’Amour, J. et al. LTR retrotransposons transcribed in oocytes drive species-specific and heritable changes in DNA methylation. Nat. Commun. 9, 3331 (2018).
Percharde, M. et al. A LINE1–Nucleolin partnership regulates early development and ESC identity. Cell 174, 391– 405 (2018).
Zalzman, M. et al. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature 464, 858–863 (2010).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Ramírez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protocols 7, 562–578 (2012).
Xi, Y. & Li, W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics 10, 232 (2009).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Schug, J. et al. Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol. 6, R33 (2005).
Dennis, G., Jr et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 4, 3 (2003).
McLean, C. Y. et al. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010).
Liu, S. et al. From 1D sequence to 3D chromatin dynamics and cellular functions: a phase separation perspective. Nucleic Acids Res. 46, 9367–9383 (2018).
Acknowledgements
We thank members of the Xie laboratory for discussions and the comments; the Animal Center and Biocomputing Facility at Tsinghua University for support; S. Gao and X. Kou for B6D2F1/J mice; and X. Shen, T. Li and X. Ji for reagents. This work was supported by the National Natural Science Foundation of China (31988101 to W. Xie), the National Key R&D Program of China (2019YFA0508900 to W. Xie and Y.Y., 2019YFA0110001 and 2017YFA0102802 to J. Na), the National Natural Science Foundation of China (31830047, 31725018 to W. Xie and 31970819 to J. Na), the Tsinghua-Peking Center for Life Sciences (W. Xie), and the Beijing Municipal Science and Technology Commission (grant Z181100001318006 to W. Xie). B.L. is supported by postdoctoral fellowships from the Tsinghua Shuimu Scholar. Q.X. and Q.W. are supported by postdoctoral fellowships from the Tsinghua-Peking Center for Life Sciences. W. Xie is an HHMI international research scholar.
Author information
Authors and Affiliations
Contributions
B.L., Q.X., Q.W. and W. Xie conceived and designed the project. B.L. developed Stacc–seq with help from W. Xia, C.Q., J.W., G.Y. and P.W., and performed Stacc–seq experiments. S.F., F.Z., J. Nie, C.Q. and B.L. designed and prepared the pA–Tn5 and pG–Tn5 fusion proteins. Q.X. and Q.W. collected and prepared the oocytes and embryos with help from Y.X., F.L., L. Liu and C.H. Y.X. collected mouse tissue samples. Q.W. and B.L. prepared mESC samples. F.L. prepared parthenogenetically activated embryos. Q.X. performed the mouse embryo experiments and immunostaining. Q.W. performed RNA-seq experiments. Y.Y. helped with the initial Stacc–seq design discussion. P.W., J. Na, L. Li, Z.L., K.X., Z.X. and F.K. helped with various experiments. B.L. analysed data with help from J.W. B.L., Q.X., Q.W. and W. Xie interpreted the data and prepared the figures. Q.X. and W. Xie wrote the manuscript with help from B.L., Q.W. and the remaining authors.
Corresponding author
Ethics declarations
Competing interests
S.F., F.Z., J. Nie, and C.Q. are employees of Vazyme Inc. The other authors declare no competing interests.
Additional information
Peer review information Nature thanks Samantha A. Morris, Keji Zhao and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Comparison of Stacc–seq, CUT&Tag and CoBATCH on histone mark profiling.
a, UCSC browser views showing the H3K4me3- and H3K27me3 enrichment detected by Stacc–seq (two biological replicates), CUT&Tag (this study; two biological replicates) and CoBATCH18. IgG or no antibody controls for Stacc–seq and the ENCODE ChIP-seq references47 are also shown. b, Bar charts showing the percentages of H3K4me3 and H3K27me3 ChIP-seq total/promoter peaks48 that are recaptured by Stacc–seq, CUT&Tag and CoBATCH. c, Box plots showing H3K4me3 and H3K27me3 enrichment by Stacc–seq, CUT&Tag and CoBATCH with various numbers of cells at peaks detected by ChIP-seq. The median is indicated by the centre line. The bottom, top edges and whiskers represent the 25th and 75th percentiles and 1.5 times the interquartile range (IQR), respectively. d, Receiver operating characteristics (ROC) curves of H3K4me3- and H3K27me3 Stacc–seq (two biological replicates), CUT&Tag (this study; two biological replicates) and CoBATCH18 with various numbers of mESCs. A published H3K27me3 CUT&Tag data from K562 cells16 were similarly analysed for comparison. Standards of ROC are the peaks from ENCODE H3K4me3 and H3K27me3 ChIP-seq data48.
Extended Data Fig. 2 Validation of Pol II Stacc–seq data in mESCs and tissues.
a, UCSC browser views showing Pol II enrichment detected by Stacc–seq using mESCs with cell number ranging from 106 to 500 (two biological replicates) and ENCODE ChIP-seq data48. A different Pol II antibody (ab2) was also applied using 500 mESCs (Methods). b, Bar charts showing the Pearson correlation coefficients between replicates of Pol II Stacc–seq (two biological replicates) using various numbers of mESCs. c, Bar charts showing the percentages of Pol II total or promoter plus gene body ChIP-seq peaks in mESC recaptured by Stacc–seq using various numbers of mESCs. d, Box plots showing the Pol II ChIP-seq enrichment48 that are recaptured or missed by Stacc–seq (two biological replicates) using various numbers of mESCs. The median is indicated by the centre line. The bottom, top edges and whiskers represent the 25th and 75th percentiles and 1.5 times the interquartile range (IQR), respectively. e, UCSC browser views showing the Stacc–seq and ENCODE ChIP-seq signals48 of Pol II enrichment in multiple mouse tissues (n = 1 biologically experiment). f, The average Pol II enrichment (spike-in normalized) at all TSSs in various numbers of mESCs in three independent batches of experiments.
Extended Data Fig. 3 Validation of Pol II Stacc–seq data in oocytes.
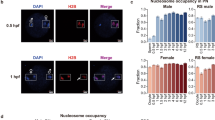
a, Immunostaining of Pol II (green) and DAPI (blue) in FGO-N and FGO-SN with (FGO-N, n = 15; FGO-SN, n = 16) or without (FGO-N, n = 18; FGO-SN, n = 15) pre-extraction21. One representative image from 3 independent experiments is shown. Dashed squares represent the location of nucleus. Scale bar: 20 μm. Quantification of Pol II intensity and P values (t-test, two-sided) are shown (right), with mean value indicated by horizontal lines. Error bar represents s.e.m. Dash line indicates the background level. b, Immunostaining of Pol II (green) at individual developmental stage. FGO-N (n = 18); FGO-SN (n = 15); MII (n = 6), PN0 one-cell (n = 6); PN2 one-cell (n = 6); PN3 one-cell (n = 15); PN5 one-cell (n = 5); 2C (n = 8), 2-cell; 8C (n = 5), 8-cell, ICM (n = 5) from blastocysts. One representative image from 3 independent experiments is shown. Dashed square represents the location of nucleus. Scale bar: 20 μm. c, UCSC browser views showing spike-in normalized Pol II enrichment in FGO-N and FGO-SN samples (two biological replicates). d, Line charts showing the average Pol II enrichment (spike-in normalized) at all TSSs after spike-in normalization in FGO-N and FGO-SN samples in two replicates. e, Bar charts showing the purity of FGO-N and FGO-SN samples from two independent experiments. A subset of oocytes from the same population used for Pol II Stacc–seq were subjected to DAPI staining, and the total numbers of sampled oocytes and the percentages of true FGO-N and FGO-SN cells (based on the absence or presence of surrounded nucleolus) in each sample are shown. The residual Pol II signals in FGO-SN samples may stem from the small subset of contaminating FGO-N cells.
Extended Data Fig. 4 Validation of Stacc–seq of Pol II in oocytes and embryos.
a, Correlation heat maps (Pearson correlation) comparing the Pol II Stacc–seq signals between replicates among stages (two biological replicates). b, Hierarchical clustering of Pol II and open chromatin in oocytes and embryos based on their global enrichment. P, paternal; M, maternal. c, Heat maps showing the Pol II, open chromatin24,25 and H3K36me338 enrichment (Z-score normalized, Methods) at shared, Pol II-specific and open chromatin-specific peaks in FGOs, early embryos and mESCs. d, UCSC browser views showing spike-in normalized Pol II- and IgG control signals in FGO-N and one-cell PN5 embryos (two biological replicates).
Extended Data Fig. 5 Pol II dynamics from one-cell to late 2-cell stages.
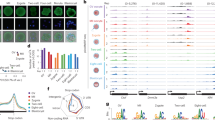
a, Heat maps showing expression levels of poising regulators in embryos, mESCs and tissues. b, Immunostaining of Cyclin T1 (1C, n = 10; Early 2C, n = 10; Late 2C, n = 20), NELFB (1C, n = 9; Early 2C, n = 12; Late 2C, n = 15) and NELFE (1C, n = 11; Early 2C, n = 8; Late 2C, n = 9) at individual stages. One representative image from 3 independent experiments is shown. Scale bar: 20 μm. c, A pie chart showing Pol II peak distribution at promoter, gene body and intergenic regions in one-cell embryos. The total number of genes covered by promoter or gene body Pol II is shown. d, Venn diagram showing the identified minor ZGA genes from total RNA-seq (this study) at one-cell and early 2-cell stages (Methods). e, Schematic illustration of α-amanitin treatment administrated at various time points. f, EU (red) staining in control and α-amanitin treated one-cell embryos (control, n = 6; α-amanitin, n = 19) (left). Immunostaining of Pol II (green) in embryos treated either with or without α-amanitin. one-cell (1C) (control, n = 13; α-amanitin, n = 15), and late 2-cell (Late 2C) (control, n = 29; α-amanitin, n = 24). One representative image from 3 independent experiments is shown. Dashed square represents the location of nucleus. Dashed circle represents parental pronucleus. Scale bar: 20 μm. Quantification of Pol II intensity and P values (t-test, two-sided) are shown (right), with mean value indicated by horizontal lines. Error bar represents s.e.m. g, UCSC browser views showing Pol II enrichment in control and α-amanitin treated one-cell (blue), one-cell MII-PN5 (purple) and late 2-cell (green) embryos before and after spike-in normalization (two biological replicates).
Extended Data Fig. 6 Pol II elongation activity in early embryos.
a, UCSC views showing Pol II enrichment (two biological replicates) at various DNA elements. b, UCSC views showing Pol II enrichment either with or without DRB treatment (two biological replicates) before and after spike-in normalization. c, Average Pol II enrichment (Z-score normalized, Methods) at MERVL and top 1,000 genes (with highest average gene body Pol II levels for each stage in control) in the presence or absence of DRB. The upstream and downstream genes are 5 × MERVL length away from the MERVL, or 1 × gene body length away from the gene. Due to DRB induced Pol II reduction, gene body Pol II enrichment at top 1,000 genes was normalized by setting the background level (far end of TSS upstream) of all samples as 0, and promoter peaks as 1. d, EU staining of control (n = 10) and DRB (n = 10) treated PN5 embryos (left), as well as immunostaining of Pol II in PN5 (control, n = 13; DRB, n = 10), early 2-cell (control, n = 10; DRB, n = 12) and late 2-cell (control, n = 14; DRB, n = 14) are shown. One representative image from 3 independent experiments is shown. Dashed square represents the nucleus. Scale bar: 20 μm. Quantification of Pol II intensity and P values (t-test, two-sided) are shown (right), with mean value indicated by horizontal lines. Error bar represents s.e.m. e, UCSC views showing Pol II enrichment (two biological replicates) at individual stages. Domains of Promiscuous transcription (PT) in one-cell were identified by total RNA10. f, Line charts showing average Pol II enrichment (Z-score normalized, Methods) and RNA levels in PT domains. The box plots show that the total RNA signals in PT domains are low compared to typical transcription signals (exons of ZGA genes as examples). The median is indicated by the centre lines. The bottom, top edges and whiskers represent the 25th and 75th percentiles and 1.5 times the IQR, respectively.
Extended Data Fig. 7 Dynamics of allelic Pol II.
a, UCSC views showing allelic Pol II on chr18 and chrX (two biological replicates). b, Box plots showing the differences of read numbers of Pol II peaks between two alleles (paternal - maternal) divided by the total number of allelic reads (paternal + maternal) (covered by at least 20 reads) on all chromosomes, chr18 and chrX at individual developmental stages. P values (t-test, one-sided, n (8-cell chr18 peaks) = 500, n (8-cell chrX peaks) = 530, n (ICM chr18 peaks) = 691, n (ICM chrX peaks) = 500) are shown. The median is indicated by the centre line. The bottom, top edges and whiskers represent the 25th and 75th percentiles and 1.5 times the interquartile range (IQR), respectively. c, Heat maps showing gene expression (FPKM) and allelic Pol II enrichment (Z-score normalized, Methods) at promoters and gene bodies on chrX of ICM, ranked by FPKM on each allele. d, UCSC browser views showing allelic Pol II enrichment and expression of Xist (two biological replicates). e, UCSC browser views showing the allelic Pol II enrichment in FGO-N and embryos (two biological replicates). To be comparable, only regions covered by SNPs in early embryos are shown for FGO-N and sperm Pol II Ser5-P ChIP-seq data34. M, maternal allele; P, paternal allele. f, Gene ontology and GREAT analysis for shared (n = 974), maternal-specific (n = 194) and paternal-specific promoters (n = 259) or distal shared (n = 13,534), maternal-specific (n = 26,528) and paternal-specific (n = 33,768) peaks in one-cell embryos. g, UCSC browser views showing the allelic enrichment of DNA methylation36, Pol II (two biological replicates), DHS24, H3K4me335 and H3K27me328 at one-cell stage at gene dense (CGI-rich) and gene poor (CGI-poor) regions64. Maternal- and paternal-specific Pol II are shaded by pink or blue, respectively. CGI, CpG islands.
Extended Data Fig. 8 Pol II enrichment and epigenetic states near DNA methylation- or H3K27me3-dependent imprinted genes.
a, Heat maps showing the levels of DNA methylation (mC), Pol II, H3K9me3 and open chromatin (ATAC-seq/DHS-seq) at DNA methylation-dependent imprinting control regions. mC and Pol II enrichment in Dnmt3l−/− oocytes or one-cell parthenogenetic (PG) embryos are also shown. b, UCSC browser views showing the allelic Pol II enrichment and mC level (two biological replicates) at ICRs in WT oocytes or embryos or Dnmt3l−/− parthenogenetic one-cell embryos. Heat maps showing the allelic gene expression levels. Kdm5a is a non-imprinted gene as control. c, Box plots and heat maps showing the levels of H3K27me328, Pol II and open chromatin25 at promoters of H3K27me3-dependent imprinted genes24 at individual developmental stages. P values (t-test, one-sided, sample n = 28) are shown. The median is indicated by the centre line. The bottom, top edges and whiskers represent the 25th and 75th percentiles and 1.5 times the interquartile range (IQR), respectively. Open circles indicate insufficient SNP coverages.
Extended Data Fig. 9 Pol II enrichment at PMD and FMD.
a, Heat maps comparing the allelic Pol II enrichment (Z-score normalized, Methods) in one-cell embryos at shared, maternal-specific and paternal-specific one-cell peaks. Pol II enrichment in one-cell parthenogenetic embryos is mapped. b, Schematic illustration of FMD, H3K4me3- and H3K27me3-marked PMD in FGOs (top). The average H3K4me335, H3K27me328, DHS24 and Pol II enrichment (Z-score normalized, Methods) are shown at FMD (bottom, left), H3K4me3-marked PMD (H3K4me3-PMD) and H3K27me3-marked PMD (H3K27me3-PMD) (bottom, right)28 between gene dense (CGI-rich) and gene poor (CGI-poor) regions in oocytes and one-cell embryos. c, UCSC browser views showing mC levels and Pol II enrichment (two biological replicates) in oocytes, WT and Dnmt3l−/− one-cell parthenogenetic embryos. Regions showing Pol II enrichment in PMD in WT but not in mutant embryos (light pink), or Pol II enrichment in FMD in mutant embryos but not in WT embryos (pale blue) are shaded.
Extended Data Fig. 10 Pol II pre-configuration before major ZGA.
a, Box plots showing the average Pol II enrichment (Z-score normalized, Methods) of oocyte-specific genes (n = 1,020), major ZGA genes (n = 438) and developmental genes (n = 3,330) at one-cell, early 2-cell and late 2-cell stages. The median is indicated by the centre line. The bottom, top edges and whiskers represent the 25th and 75th percentiles and 1.5 times the interquartile range (IQR), respectively. b, Heat maps showing enrichment of all repeat subfamilies at Pol II promoter (left) and distal (right) peaks (compared to that in random peaks) in oocytes, early embryos and mESCs (numbers of promoter peaks from FGO-N to mESC are 8,072, 18,139, 16,523, 12,411, 13,537, 12,934, 14,747, respectively; numbers of distal peaks from FGO-N to mESC are 51,342, 55,985, 51,173, 36,640, 45,456, 31,737, 27,094, respectively). Pol II enrichment was calculated as a log2 ratio of the numbers of observed peaks that overlap with repeats divided by the numbers of random peaks that overlap with repeats. c, Heat maps showing Pol II and H3K4me335 levels from various stages at shared, one-cell-specific and late 2-cell-specific Pol II peaks for each allele. The upstream and downstream regions are 1 × peak length away from the H3K4me335 enrichment at Pol II one-cell and late 2-cell-specific peaks. d, Hierarchical clustering of allelic H3K4me335 at Pol II one-cell-specific and late 2-cell-specific peaks.
Extended Data Fig. 11 Pol II pre-configuration depends on minor ZGA.
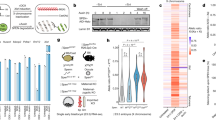
a, Line charts showing the average Pol II enrichment at all PN5 one-cell (left) and early 2-cell (right) peaks in PN5 one-cell, mitotic one-cell embryos in the presence or absence of transcription (TBE) (two biological replicates), and early 2-cell embryos. b, UCSC browser views showing Pol II enrichment in samples described in a. c, Developmental rate of minor-ZGA transcription blocked embryos (mTBEs). Various concentrations of DRB were used. d, WNT3A (mTBE-specific gene) (control, n = 20; mTBE, n = 20) and NR5A2 (ZGA gene) (control, n = 13; mTBE, n = 20) staining in control and mTBE embryos. Scale bar: 20 μm. Quantification of IF signal and P values (t-test, two-sided) are shown (right), with mean value indicated by horizontal lines. Error bar represents s.e.m. One representative image from 3 independent experiments is shown. e, Pol II staining in control and mTBE embryos (control before wash, n = 32; DRB before wash, n = 10; control after wash, n = 20; DRB after wash; n = 38) (left). One representative image from 3 independent experiments is shown. Dashed square represents the location of nucleus. Scale bar: 20 μm. Quantification of Pol II signal and P values (t-test, two-sided) are shown (right), with mean value indicated by horizontal lines. Error bar represents s.e.m.
Extended Data Fig. 12 The effects of DRB on Pol I and Pol II transcription in embryos.
a, Box plots showing top 100 major ZGA and mTBE-specific gene expression (quantile normalized FPKM) in this study and previous studies10,15 and gene body Pol II enrichment (Z-score normalized, Methods; two biological replicates). Major ZGA top 100 genes were selected by ZGA genes with highest Pol II enrichment increase (late 2-cell–one-cell) on the gene bodies. P values (t-test, two-sided, top 100 ZGA genes, sample size n = 100; and mTBE-specific gene, sample size n = 481) are shown. The median is indicated by the centre lines. The bottom, top edges and whiskers represent the 25th and 75th percentiles and 1.5 times the IQR, respectively. Gene ontology results for the top 100 major ZGA and mTBE specific genes are shown (right). b, qPCR analyses of rRNA and minor ZGA gene (Zfp352 and Zscan4) expression in DMSO- and DRB-treated early 2-cell embryos (two biological replicates). Gene expression was first normalized using either the expression of Gapdh or the absolute cell numbers, and the DRB/DMSO ratios were then computed. Data are presented as mean values. Dots indicate each data point.
Supplementary information
Supplementary Table
Supplementary Table 1: Sample information. The sample information includes source name, developmental stage, cell number, experiment type, spike-in DNA, treatment, genotype and antibody for all samples.
Supplementary Table
Supplementary Table 2: Genes ectopically expressed in mTBEs. Genes ectopically expressed in mTBEs (minor-ZGA blocked embryos) are included.
Rights and permissions
About this article
Cite this article
Liu, B., Xu, Q., Wang, Q. et al. The landscape of RNA Pol II binding reveals a stepwise transition during ZGA. Nature 587, 139–144 (2020). https://doi.org/10.1038/s41586-020-2847-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2847-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.