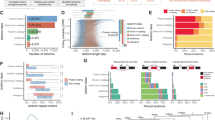
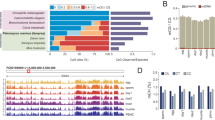
Abstract
Single-cell RNA sequencing of embryos can resolve the transcriptional landscape of development at unprecedented resolution. To date, single-cell RNA-sequencing studies of mammalian embryos have focused exclusively on eutherian species. Analysis of mammalian outgroups has the potential to identify deeply conserved lineage specification and pluripotency factors, and can extend our understanding of X dosage compensation. Metatherian (marsupial) mammals diverged from eutherians around 160 million years ago. They exhibit distinctive developmental features, including late implantation1 and imprinted X chromosome inactivation2, which is associated with expression of the XIST-like noncoding RNA RSX3. Here we perform a single-cell RNA-sequencing analysis of embryogenesis and X chromosome inactivation in a marsupial, the grey short-tailed opossum (Monodelphis domestica). We resolve the developmental trajectory and transcriptional signatures of the epiblast, primitive endoderm and trophectoderm, and identify deeply conserved lineage-specific markers that pre-date the eutherian–marsupial divergence. RSX coating and inactivation of the X chromosome occurs early and rapidly. This observation supports the hypothesis that—in organisms with early X chromosome inactivation—imprinted X chromosome inactivation prevents biallelic X silencing. We identify XSR, an RSX antisense transcript expressed from the active X chromosome, as a candidate for the regulator of imprinted X chromosome inactivation. Our datasets provide insights into the evolution of mammalian embryogenesis and X dosage compensation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All cytological data and R markdown files in this study are included in this published Article, and its Supplementary Information (Supplementary Information 3–6). Sequence data have been deposited at ArrayExpress (accession code E-MTAB-7515).
Change history
16 October 2020
A Correction to this paper has been published: https://doi.org/10.1038/s41586-020-2840-5
References
Frankenberg, S. R., de Barros, F. R., Rossant, J. & Renfree, M. B. The mammalian blastocyst. Wiley Interdiscip. Rev. Dev. Biol. 5, 210–232 (2016).
Sharman, G. B. Late DNA replication in the paternally derived X chromosome of female kangaroos. Nature 230, 231–232 (1971).
Grant, J. et al. Rsx is a metatherian RNA with Xist-like properties in X-chromosome inactivation. Nature 487, 254–258 (2012).
Graves, J. A. & Renfree, M. B. Marsupials in the age of genomics. Annu. Rev. Genomics Hum. Genet. 14, 393–420 (2013).
Samollow, P. B. The opossum genome: insights and opportunities from an alternative mammal. Genome Res. 18, 1199–1215 (2008).
Murchison, E. P. et al. Genome sequencing and analysis of the Tasmanian devil and its transmissible cancer. Cell 148, 780–791 (2012).
VandeBerg, J. L., Williams-Blangero, S., Hubbard, G. B., Ley, R. D. & Robinson, E. S. Genetic analysis of ultraviolet radiation-induced skin hyperplasia and neoplasia in a laboratory marsupial model (Monodelphis domestica). Arch. Dermatol. Res. 286, 12–17 (1994).
Selwood, L. Mechanisms underlying the development of pattern in marsupial embryos. Curr. Top. Dev. Biol. 27, 175–233 (1992).
Frankenberg, S., Shaw, G., Freyer, C., Pask, A. J. & Renfree, M. B. Early cell lineage specification in a marsupial: a case for diverse mechanisms among mammals. Development 140, 965–975 (2013).
Morrison, J. T., Bantilan, N. S., Wang, V. N., Nellett, K. M. & Cruz, Y. P. Expression patterns of Oct4, Cdx2, Tead4, and Yap1 proteins during blastocyst formation in embryos of the marsupial, Monodelphis domestica Wagner. Evol. Dev. 15, 171–185 (2013).
Deng, X., Berletch, J. B., Nguyen, D. K. & Disteche, C. M. X chromosome regulation: diverse patterns in development, tissues and disease. Nat. Rev. Genet. 15, 367–378 (2014).
Okamoto, I. et al. Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature 472, 370–374 (2011).
Mak, W. et al. Reactivation of the paternal X chromosome in early mouse embryos. Science 303, 666–669 (2004).
Okamoto, I., Otte, A. P., Allis, C. D., Reinberg, D. & Heard, E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science 303, 644–649 (2004).
Duret, L., Chureau, C., Samain, S., Weissenbach, J. & Avner, P. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science 312, 1653–1655 (2006).
Petropoulos, S. et al. Single-cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell 165, 1012–1026 (2016).
Chambers, I. et al. Nanog safeguards pluripotency and mediates germline development. Nature 450, 1230–1234 (2007).
Mitsui, K. et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113, 631–642 (2003).
Chia, N. Y. et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature 468, 316–320 (2010).
Yamaji, M. et al. PRDM14 ensures naive pluripotency through dual regulation of signaling and epigenetic pathways in mouse embryonic stem cells. Cell Stem Cell 12, 368–382 (2013).
Nichols, J. et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95, 379–391 (1998).
Fogarty, N. M. E. et al. Genome editing reveals a role for OCT4 in human embryogenesis. Nature 550, 67–73 (2017).
Frankenberg, S., Pask, A. & Renfree, M. B. The evolution of class V POU domain transcription factors in vertebrates and their characterisation in a marsupial. Dev. Biol. 337, 162–170 (2010).
Koutsourakis, M., Langeveld, A., Patient, R., Beddington, R. & Grosveld, F. The transcription factor GATA6 is essential for early extraembryonic development. Development 126, 723–732 (1999).
Morrisey, E. E. et al. GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes Dev. 12, 3579–3590 (1998).
Wamaitha, S. E. et al. Gata6 potently initiates reprograming of pluripotent and differentiated cells to extraembryonic endoderm stem cells. Genes Dev. 29, 1239–1255 (2015).
Shimosato, D., Shiki, M. & Niwa, H. Extra-embryonic endoderm cells derived from ES cells induced by GATA factors acquire the character of XEN cells. BMC Dev. Biol. 7, 80 (2007).
Fujikura, J. et al. Differentiation of embryonic stem cells is induced by GATA factors. Genes Dev. 16, 784–789 (2002).
Mate, K. E., Robinson, E. S., Vandeberg, J. L. & Pedersen, R. A. Timetable of in vivo embryonic development in the grey short-tailed opossum (Monodelphis domestica). Mol. Reprod. Dev. 39, 365–374 (1994).
Kang, M., Garg, V. & Hadjantonakis, A. K. Lineage establishment and progression within the inner cell mass of the mouse blastocyst requires FGFR1 and FGFR2. Dev. Cell 41, 496–510 (2017).
Molotkov, A., Mazot, P., Brewer, J. R., Cinalli, R. M. & Soriano, P. Distinct requirements for FGFR1 and FGFR2 in primitive endoderm development and exit from pluripotency. Dev. Cell 41, 511–526 (2017).
Rossant, J. Genetic control of early cell lineages in the mammalian embryo. Annu. Rev. Genet. 52, 185–201 (2018).
Morris, S. A., Graham, S. J., Jedrusik, A. & Zernicka-Goetz, M. The differential response to Fgf signalling in cells internalized at different times influences lineage segregation in preimplantation mouse embryos. Open Biol. 3, 130104 (2013).
Artus, J., Panthier, J. J. & Hadjantonakis, A. K. A role for PDGF signaling in expansion of the extra-embryonic endoderm lineage of the mouse blastocyst. Development 137, 3361–3372 (2010).
Yagi, R. et al. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development 134, 3827–3836 (2007).
Nishioka, N. et al. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech. Dev. 125, 270–283 (2008).
Krendl, C. et al. GATA2/3-TFAP2A/C transcription factor network couples human pluripotent stem cell differentiation to trophectoderm with repression of pluripotency. Proc. Natl Acad. Sci. USA 114, E9579–E9588 (2017).
Strumpf, D. et al. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 132, 2093–2102 (2005).
Wu, G. et al. Initiation of trophectoderm lineage specification in mouse embryos is independent of Cdx2. Development 137, 4159–4169 (2010).
Blakeley, P. et al. Defining the three cell lineages of the human blastocyst by single-cell RNA-seq. Development 142, 3613 (2015).
Nakamura, T. et al. A developmental coordinate of pluripotency among mice, monkeys and humans. Nature 537, 57–62 (2016).
Nakamura, T. et al. SC3-seq: a method for highly parallel and quantitative measurement of single-cell gene expression. Nucleic Acids Res. 43, e60 (2015).
Mahadevaiah, S. K. et al. Key features of the X inactivation process are conserved between marsupials and eutherians. Curr. Biol. 19, 1478–1484 (2009).
Wang, X., Douglas, K. C., Vandeberg, J. L., Clark, A. G. & Samollow, P. B. Chromosome-wide profiling of X-chromosome inactivation and epigenetic states in fetal brain and placenta of the opossum, Monodelphis domestica. Genome Res. 24, 70–83 (2014).
Sahakyan, A. et al. Human naive pluripotent stem cells model X chromosome dampening and X inactivation. Cell Stem Cell 20, 87–101 (2017).
Duthie, S. M. et al. Xist RNA exhibits a banded localization on the inactive X chromosome and is excluded from autosomal material in cis. Hum. Mol. Genet. 8, 195–204 (1999).
Inoue, A., Jiang, L., Lu, F. & Zhang, Y. Genomic imprinting of Xist by maternal H3K27me3. Genes Dev. 31, 1927–1932 (2017).
Lee, J. T. Disruption of imprinted X inactivation by parent-of-origin effects at Tsix. Cell 103, 17–27 (2000).
Maclary, E. et al. Differentiation-dependent requirement of Tsix long non-coding RNA in imprinted X-chromosome inactivation. Nat. Commun. 5, 4209 (2014).
Sado, T., Wang, Z., Sasaki, H. & Li, E. Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development 128, 1275–1286 (2001).
Ohhata, T., Senner, C. E., Hemberger, M. & Wutz, A. Lineage-specific function of the noncoding Tsix RNA for Xist repression and Xi reactivation in mice. Genes Dev. 25, 1702–1715 (2011).
Rossant, J., Genetic control of early cell lineages in the mammalian embryo. Ann. Rev. Genet. 52, 185–201 (2018).
Hadfield, J. & Eldridge, M. D. Multi-genome alignment for quality control and contamination screening of next-generation sequencing data. Front. Genet. 5, 31 (2014).
Cortez, D. et al. Origins and functional evolution of Y chromosomes across mammals. Nature 508, 488–493 (2014).
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T. & Salzberg, S. L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protocols 11, 1650–1667 (2016).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Qiu, X. et al. Single-cell mRNA quantification and differential analysis with Census. Nat. Methods 14, 309–315 (2017).
Turner, J. M., Mahadevaiah, S. K., Ellis, P. J., Mitchell, M. J. & Burgoyne, P. S. Pachytene asynapsis drives meiotic sex chromosome inactivation and leads to substantial postmeiotic repression in spermatids. Dev. Cell 10, 521–529 (2006).
Sangrithi, M. N. et al. Non-canonical and sexually dimorphic X dosage compensation states in the mouse and human germline. Dev. Cell 40, 289–301 (2017).
Acknowledgements
This work was supported by European Research Council (CoG 647971) and the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001193), UK Medical Research Council (FC001193) and Wellcome Trust (FC001193). We thank the Francis Crick Institute Biological Research, Advanced Sequencing and R. Goldstone, and Light Microscopy facilities and D. Bell for their expertise; H. Niwa for the POU5F3 antibody; D. Page for the MAB1980 (opossum Y chromosome) probe; and members of the J.M.A.T. and K. Niakan laboratories for comments on the manuscript.
Author information
Authors and Affiliations
Contributions
J.M.A.T. and S.K.M. conceived the project. S.K.M. performed and optimized embryo collection, single-cell collection, RNA extraction, cDNA synthesis, RNA FISH, DNA FISH, immunofluorescence, RT–PCR and MiSeq. M.N.S. performed computational analysis. T.H. advised on data representation and generated figures. J.M.A.T., S.K.M. and M.N.S. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Takashi Sado and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Illustration of late opossum embryo development and single-cell clustering.
a, Schematic of opossum embryos at E8–E12.5. b, Single-cell correlation coefficients. c, Principal component analysis.
Extended Data Fig. 2 Embryonic genome activation, Y gene expression and clustering by unsupervised 3D t-SNE.
a, Y gene expression in scRNA-seq data derived from oocytes (n = 6 cells) and presumptive male embryos at E1.5 (n = 7 embryos) and E2.5 (n = 5 embryos), and E3.5 to E7.5 (n = 20 embryos). Y transcripts cannot be maternally deposited, because the Y chromosome is inherited from the father. Their appearance must therefore signify the initiation of embryonic transcription. b, Immature and mature (asterisk) nucleolar morphology in embryos at E2.5 and E3.5, respectively. Images are maximum projections. Scale bars, 10 μm. c, Percentage of multi-allelic X- and autosomally encoded SNP variants in male bulk RNA-seq data (n = 28 embryos). When multiple SNPs for X-encoded RNAs are detected, maternal RNA is still present. However, when single SNPs are detected, it has been degraded. d, Unsupervised clustering of scRNA-seq data. e, Gene ontology analysis for genes in d.
Extended Data Fig. 3 Identification of lineage-specific genes.
a, Pseudotemporal ordering from Fig. 2c in 3D. b, Pseudotemporal ordering from Fig. 2c separated by sex. c, Pseudotemporal ordering from Fig. 2c, with cells coloured according to developmental age. d, Full heat map of genes defining cluster 1, cluster 2, early EPI, EPI, PrE, early TE and TE. e, Gene ontology analysis for cluster 1 and cluster 2, early EPI and early TE.
Extended Data Fig. 4 Lineage marker gene expression and immunostaining.
a, Expression of representative genes in cluster 1, early EPI, EPI, PrE, early TE and TE. b, Expression of GATA6 and TEAD4 at E5.5. All cells are TEAD4-negative, even those that are not expressing GATA6 (encircled). Images are maximum projections. Scale bar, 10 μm.
Extended Data Fig. 5 Lineage clusters in eutherians.
a–c, Semi-supervised 3D t-SNE (left) and heat map (right) of mouse (a), macaque (b) and human (c) embryos, with single cells coloured according to cluster.
Extended Data Fig. 6 Eutherian and marsupial lineage conservation.
a, Genes expressed in EPI, PrE and TE in mouse, macaque and human. b, Heat map showing expression of conserved genes in opossum. c, Expression of opossum genes, the orthologues of which are associated with naive and primed pluripotency in macaque. d, Expression heat map of opossum genes with mouse orthologues that are expressed specifically in mouse EPI at E6.5 (Coro1a) or E4.5 (all other genes).
Extended Data Fig. 7 Analysis of X inactivation.
a, RSX expression in female opossums. b, Dual RSX and MSN RNA FISH in embryos at E2.5 (16 embryos = 68 cells). c, Dual RSX and MSN RNA FISH (left), followed by Y chromosome DNA FISH (right) in male embryos at E4.5; blastomeres magnified in insets. d, Y gene versus RSX expression. e, MSN RNA FISH followed by Y chromosome DNA FISH in opossum spermatids. f, Dual RSX and MSN RNA FISH in female opossum brain cells. Images are maximum projections. Scale bars, 10 μm. g, Percentage of multi-allelic X SNP variants in female (n = 15 embryos) and male (n = 28 embryos) bulk RNA-seq data from E3.5 to E7.5. Mann–Whitney U-test was used to calculate P values. For RNA FISH data, quantification is shown below the images. h, SNP analysis showing expression of 11 genes from the paternal X chromosome in embryos at E3.5 (n = 3) and E4.5 (n = 2). The location of RSX is shown. Asterisks denote instances in which informative SNPs were not present. ATRX is the only gene that exhibits no expression from the paternal X chromosome at E3.5 (star): expression from the maternal X chromosome at this age could represent maternal products, with activation of embryonic ATRX initiating at E4.5.
Extended Data Fig. 8 Further analysis of X inactivation.
a, RSX localization to the inactive X chromosome (identified using MSN DNA FISH) in dividing cells. Images are maximum projections. Scale bar, 10 μm. b, X chromosome-to-autosome ratios in female clusters at E6.0–E7.5. c, Reverse transcription PCR analysis of RSX, XSR, TFE3Y and GAPDH in female and male embryos at E5.5. d, RSX and XSR expression in adult female and male tissues. e, Dual RSX and XSR RNA FISH in female and male embryos at E3.5. The explanation for why the RSX and XSR FISH signals are the same colour is given in the Methods.
Supplementary information
Supplementary Information 9
Supplementary discussion. Interrogation of the pluriblast cell population in opossum embryos.
Supplementary Table 1
. Differential expression test results of lineage markers. Relates to Fig.2a, b, d and Extended Data Figure 3.
Supplementary Table 2
. Eutherian - marsupial lineage conservation analysis. Relates to Extended Data Figure 6.
Supplementary Information 3
. R markdown html file for Monodelphis domestica analysis.
Supplementary Information 4
. R markdown html file for Macaca fascicularis analysis.
Supplementary Information 5
. R markdown html file for Homo sapiens analysis.
Supplementary Information 6
. R markdown html file for Mus musculus analysis.
Supplementary Table 7
. X chromosome genes primers. The 11 X genes primers used in identifying parental SNPs.
Supplementary Table 8
. RSX and XSR MiSeq reads with SNPs - Parent-of-origin expression of RSX and XSR in whole embryos at E3.5 and E5.5 opossum embryos.
Rights and permissions
About this article
Cite this article
Mahadevaiah, S.K., Sangrithi, M.N., Hirota, T. et al. A single-cell transcriptome atlas of marsupial embryogenesis and X inactivation. Nature 586, 612–617 (2020). https://doi.org/10.1038/s41586-020-2629-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2629-6
This article is cited by
-
Marsupials have monoallelic MEST expression with a conserved antisense lncRNA but MEST is not imprinted
Heredity (2024)
-
A hexa-species transcriptome atlas of mammalian embryogenesis delineates metabolic regulation across three different implantation modes
Nature Communications (2022)
-
Gene regulation in time and space during X-chromosome inactivation
Nature Reviews Molecular Cell Biology (2022)
-
Elastic dosage compensation by X-chromosome upregulation
Nature Communications (2022)
-
Enhanced chromatin accessibility contributes to X chromosome dosage compensation in mammals
Genome Biology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.