The COVID-19 pandemic is the biggest public-health crisis in a century, and the development of medical interventions to combat the SARS-CoV-2 coronavirus is a top priority. Writing in Nature, Pinto et al.1 provide evidence needed to take one of the crucial first steps for such efforts in the developing arena of antibody immunotherapy.
Read the paper: Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody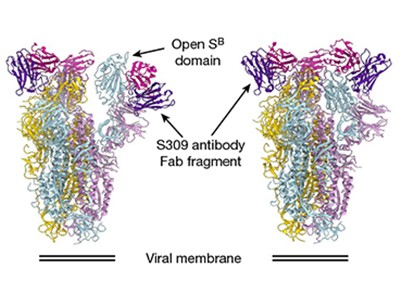
The level of protection provided by the immune system in response to SARS-CoV-2 exposure and infection is a hotly debated topic2. It is thought that one major arm of the immune response to such infection is the development of antibodies that recognize the virus. Of particular interest are antibodies that bind to a protein on the viral surface known as the spike protein. Coronaviruses derive their name from their distinctive, crown-like (coronal) viral silhouettes, which are due to these proteins.
Antibodies that recognize and bind to the viral ‘spike’ can block its ability to bind the ACE2 receptor protein on human cells. An interaction between the spike protein and ACE2 is part of a process that can enable coronaviruses to enter human cells. Thus, antibodies that could hinder spike-protein function would block infection; such antibodies are termed neutralizing antibodies.
Much remains to be learnt about the immunological responses to SARS-CoV-2. Nevertheless, it is becoming clear that antibodies taken from the blood serum of people who have recovered from COVID-19 can be used for treatment by being transfused into other people who have the disease3. Such ‘convalescent sera’ approaches are highly attractive, particularly as an immediate treatment option. That’s because more-conventional therapeutics, such as drugs or vaccines, are unlikely to be available for some time. A more high-tech approach to using convalescent sera is the manipulation of antibody-producing B cells taken from the blood of people who had COVID-19 or other coronavirus infections. Each B cell makes one unique antibody, and clonal populations of a B cell of interest can be used to generate an identical pool of a particular desired antibody known as a monoclonal antibody.
To accelerate the process of therapeutic development, Pinto and colleagues ‘went back in time’, and turned to samples of B cells collected from a person who had been infected by the coronavirus SARS-CoV. This virus, which is similar to SARS-CoV-2, caused an outbreak in 2003 of a disease called severe acute respiratory syndrome (SARS). The hope with such an approach is that the resemblance between the two viruses might mean that some antibodies that recognize SARS-CoV also recognize and neutralize SARS-CoV-2.
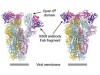
The ‘head’, or receptor-binding domain (termed S1), of the spike protein is the most accessible region of the protein for antibodies to bind to. However, this domain exists in different dynamic states, and debate has arisen over whether it is ‘masked’ from the immune system by a shell of carbohydrate molecules4. The identification of a functional antibody that targets this region is therefore not a trivial process. Pinto et al. combined blood cells taken in 2004 and 2013 from a person who had recovered from SARS, and searched for antibodies that could recognize SARS-CoV-2 (Fig. 1). Of the 25 different monoclonal antibodies that the authors studied, 4 recognized the receptor-binding domains of both SARS-CoV and SARS-CoV-2 spike proteins. One antibody, termed S309, was selected for further study on the basis of its high-affinity binding to this domain when tested in vitro.
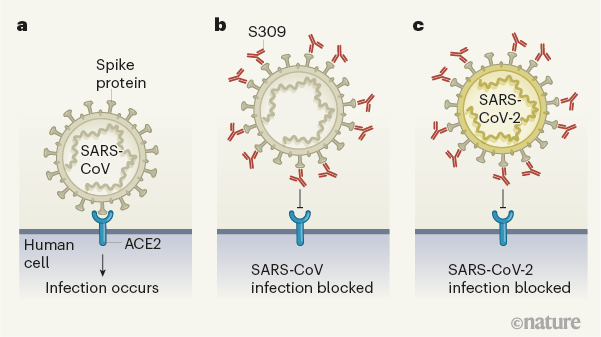
Figure 1 | An antibody that blocks coronavirus infections. Pinto et al.1 have identified a human antibody that blocks infection by SARS-CoV-2, the coronavirus that causes COVID-19. The authors made this discovery by examining antibodies made by a person who had recovered in 2003 from infection with the related coronavirus SARS-CoV, which causes severe acute respiratory syndrome (SARS). a, Coronaviruses such as SARS-CoV infect human cells by binding to the protein ACE2. b, Pinto and colleagues analysed blood samples taken in 2004 and 2011 from a person who recovered from SARS, and examined antibodies made by the immune cells from the samples. They identified an antibody (named S309) that bound to the spike protein of SARS-CoV and prevented infection by this virus. c, The authors found that this antibody bound to a similar region of the spike protein of SARS-CoV-2 and prevented infection by the virus.
Pinto and colleagues used cryo-electron microscopy to visualize the interaction between the S309 antibody and the SARS-CoV-2 spike protein. This revealed that S309 binds to an accessible site in the receptor-binding domain of the spike protein that has an attached carbohydrate molecule. This region is not part of the key area that directly binds to ACE2. The site that S309 recognizes is evolutionarily conserved in spike proteins across a range of bat coronaviruses (in the genus Betacoronavirus lineage B; subgenus Sarbecovirus) that have similarities to the SARS-like coronaviruses. This raises the possibility that such an antibody could have wide applicability in tackling related viruses. Not only, then, is this antibody of interest when investigating ways to manage the COVID-19 pandemic in the years ahead, but it might also be considered for use in preventing future outbreaks of related animal viruses, if they make the leap to causing infection in humans.
Read the paper: Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody
Ultimately, it seems unlikely that a robust treatment for COVID-19 will rely on a single antibody. Rather, as was the case for SARS, a synergistic approach combining different monoclonal antibodies in an antibody cocktail might be more effective5. For such approaches to move forwards, evidence of effective antibody neutralization from in vitro studies will be needed, along with in vivo data assessing how well an antibody can boost other aspects of the immune response — by enlisting other immune cells to tackle the infection, for example. There are many promising avenues to explore in these efforts.
Pinto and colleagues got a head start with their work by exploring pre-existing antibodies, and they should now have more B-cell populations to mine. Many other teams, to give just some examples2,6–13, have also presented useful discoveries in the hunt for antibodies that can target SARS-CoV-2. The next steps will be to test individual antibodies and antibody cocktails in animal models, to determine whether they offer protection, and then to assess their safety and effectiveness in human clinical trials. An accelerated path might narrow the time lag between antibody discovery and proof-of-concept trials in humans to as little as five or six months14.
The most recent prominent example of immunotherapy for infectious disease relates to battling the Ebola virus. In concert with vaccines and conventional, small-molecule-drug trials, the development of monoclonal-antibody therapies for Ebola has progressed rapidly. Cocktails of antibodies, beginning with one called ZMapp, that target a key Ebola viral protein called GP in two crucial regions of the protein, are continuing to be developed15–17. This progress in efforts to tackle Ebola gives hope for similar immunotherapy achievements in targeting SARS-CoV-2. Pinto and colleagues’ work marks a major step towards that much-anticipated, and much-needed, success.

 Read the paper: Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody
Read the paper: Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody
 CRISPR tool scales up to interrogate a huge line-up of viral suspects
CRISPR tool scales up to interrogate a huge line-up of viral suspects
 All for one and one for all to fight flu
All for one and one for all to fight flu