Abstract
Sex chromosomes in males of most eutherian mammals share only a small homologous segment, the pseudoautosomal region (PAR), in which the formation of double-strand breaks (DSBs), pairing and crossing over must occur for correct meiotic segregation1,2. How cells ensure that recombination occurs in the PAR is unknown. Here we present a dynamic ultrastructure of the PAR and identify controlling cis- and trans-acting factors that make the PAR the hottest segment for DSB formation in the male mouse genome. Before break formation, multiple DSB-promoting factors hyperaccumulate in the PAR, its chromosome axes elongate and the sister chromatids separate. These processes are linked to heterochromatic mo-2 minisatellite arrays, and require MEI4 and ANKRD31 proteins but not the axis components REC8 or HORMAD1. We propose that the repetitive DNA sequence of the PAR confers unique chromatin and higher-order structures that are crucial for recombination. Chromosome synapsis triggers collapse of the elongated PAR structure and, notably, oocytes can be reprogrammed to exhibit spermatocyte-like levels of DSBs in the PAR simply by delaying or preventing synapsis. Thus, the sexually dimorphic behaviour of the PAR is in part a result of kinetic differences between the sexes in a race between the maturation of the PAR structure, formation of DSBs and completion of pairing and synapsis. Our findings establish a mechanistic paradigm for the recombination of sex chromosomes during meiosis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
SSDS data are publicly available at the GEO under the accession numbers GSE35498, GSE99921 and GSE118913. Source Data for all graphs in the figures are provided with the paper (Figs. 2, 3, Extended Data Figs. 1, 3, 4, 6, 7, 9). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository44 with the dataset identifier PXD017191. Processed mass spectrometry data are provided in the Source Data for Extended Data Fig. 1.
Code availability
Image analysis scripts are available on GitHub (https://github.com/Boekhout/ImageJScripts).
References
Raudsepp, T. & Chowdhary, B. P. The eutherian pseudoautosomal region. Cytogenet. Genome Res. 147, 81–94 (2015).
Kauppi, L. et al. Distinct properties of the XY pseudoautosomal region crucial for male meiosis. Science 331, 916–920 (2011).
Perry, J., Palmer, S., Gabriel, A. & Ashworth, A. A short pseudoautosomal region in laboratory mice. Genome Res. 11, 1826–1832 (2001).
Raudsepp, T., Das, P. J., Avila, F. & Chowdhary, B. P. The pseudoautosomal region and sex chromosome aneuploidies in domestic species. Sex Dev. 6, 72–83 (2012).
Palmer, S., Perry, J., Kipling, D. & Ashworth, A. A gene spans the pseudoautosomal boundary in mice. Proc. Natl Acad. Sci. USA 94, 12030–12035 (1997).
Soriano, P. et al. High rate of recombination and double crossovers in the mouse pseudoautosomal region during male meiosis. Proc. Natl Acad. Sci. USA 84, 7218–7220 (1987).
Brick, K., Smagulova, F., Khil, P., Camerini-Otero, R. D. & Petukhova, G. V. Genetic recombination is directed away from functional genomic elements in mice. Nature 485, 642–645 (2012).
Lange, J. et al. The landscape of mouse meiotic double-strand break formation, processing, and repair. Cell 167, 695–708 (2016).
Kleckner, N. Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma 115, 175–194 (2006).
Zickler, D. & Kleckner, N. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 33, 603–754 (1999).
Panizza, S. et al. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell 146, 372–383 (2011).
Lam, I. & Keeney, S. Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb. Perspect. Biol. 7, a016634 (2014).
de Massy, B. Initiation of meiotic recombination: how and where? Conservation and specificities among eukaryotes. Annu. Rev. Genet. 47, 563–599 (2013).
Kauppi, L., Jasin, M. & Keeney, S. The tricky path to recombining X and Y chromosomes in meiosis. Ann. NY Acad. Sci. 1267, 18–23 (2012).
Page, J. et al. Inactivation or non-reactivation: what accounts better for the silence of sex chromosomes during mammalian male meiosis? Chromosoma 121, 307–326 (2012).
Kumar, R., Bourbon, H. M. & de Massy, B. Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice. Genes Dev. 24, 1266–1280 (2010).
Stanzione, M. et al. Meiotic DNA break formation requires the unsynapsed chromosome axis-binding protein IHO1 (CCDC36) in mice. Nat. Cell Biol. 18, 1208–1220 (2016).
Reinholdt, L. G. & Schimenti, J. C. Mei1 is epistatic to Dmc1 during mouse meiosis. Chromosoma 114, 127–134 (2005).
Kumar, R. et al. Mouse REC114 is essential for meiotic DNA double-strand break formation and forms a complex with MEI4. Life Sci. Alliance 1, e201800259 (2018).
Boekhout, M. et al. REC114 partner ANKRD31 controls number, timing, and location of meiotic DNA breaks. Mol. Cell 74, 1053–1068 (2019).
Papanikos, F. et al. Mouse ANKRD31 regulates spatiotemporal patterning of meiotic recombination initiation and ensures recombination between X and Y sex chromosomes. Mol. Cell 74, 1069–1085 (2019).
Kumar, R. et al. MEI4 – a central player in the regulation of meiotic DNA double-strand break formation in the mouse. J. Cell Sci. 128, 1800–1811 (2015).
Harbers, K., Francke, U., Soriano, P., Jaenisch, R. & Müller, U. Structure and chromosomal mapping of a highly polymorphic repetitive DNA sequence from the pseudoautosomal region of the mouse sex chromosomes. Cytogenet. Cell Genet. 53, 129–133 (1990).
Takahashi, Y. et al. Methylation imprinting was observed of mouse mo-2 macrosatellite on the pseudoautosomal region but not on chromosome 9. Chromosoma 103, 450–458 (1994).
Shin, Y. H. et al. Hormad1 mutation disrupts synaptonemal complex formation, recombination, and chromosome segregation in mammalian meiosis. PLoS Genet. 6, e1001190 (2010).
Bannister, L. A., Reinholdt, L. G., Munroe, R. J. & Schimenti, J. C. Positional cloning and characterization of mouse mei8, a disrupted allelle of the meiotic cohesin Rec8. Genesis 40, 184–194 (2004).
Kim, K. P. et al. Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell 143, 924–937 (2010).
Daniel, K. et al. Meiotic homologue alignment and its quality surveillance are controlled by mouse HORMAD1. Nat. Cell Biol. 13, 599–610 (2011).
Brick, K., Pratto, F., Sun, C. Y., Camerini-Otero, R. D. & Petukhova, G. Analysis of meiotic double-strand break initiation in mammals. Methods Enzymol. 601, 391–418 (2018).
Khil, P. P., Smagulova, F., Brick, K. M., Camerini-Otero, R. D. & Petukhova, G. V. Sensitive mapping of recombination hotspots using sequencing-based detection of ssDNA. Genome Res. 22, 957–965 (2012).
Brick, K. et al. Extensive sex differences at the initiation of genetic recombination. Nature 561, 338–342 (2018).
Thacker, D., Mohibullah, N., Zhu, X. & Keeney, S. Homologue engagement controls meiotic DNA break number and distribution. Nature 510, 241–246 (2014).
Kauppi, L. et al. Numerical constraints and feedback control of double-strand breaks in mouse meiosis. Genes Dev. 27, 873–886 (2013).
Arnold, A. P. Mouse models for evaluating sex chromosome effects that cause sex differences in non-gonadal tissues. J. Neuroendocrinol. 21, 377–386 (2009).
Murakami, H., Lam, I., Song, J., Huang, P.-C., Song, J., van Overbeek, M. & Keeney, S. Multilayered mechanisms ensure that short chromosomes recombine in meiosis. Nature https://doi.org/10.1038/s41586-020-2248-2 (2020).
Harbers, K., Soriano, P., Müller, U. & Jaenisch, R. High frequency of unequal recombination in pseudoautosomal region shown by proviral insertion in transgenic mouse. Nature 324, 682–685 (1986).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Kipling, D. et al. Structural variation of the pseudoautosomal region between and within inbred mouse strains. Proc. Natl Acad. Sci. USA 93, 171–175 (1996).
Kipling, D., Salido, E. C., Shapiro, L. J. & Cooke, H. J. High frequency de novo alterations in the long-range genomic structure of the mouse pseudoautosomal region. Nat. Genet. 13, 78–82 (1996).
Karasu, M. E. & Keeney, S. Cyclin B3 is dispensable for mouse spermatogenesis. Chromosoma 128, 473–487 (2019).
Karasu, M. E., Bouftas, N., Keeney, S. & Wassmann, K. Cyclin B3 promotes anaphase I onset in oocyte meiosis. J. Cell Biol. 218, 1265–1281 (2019).
R Core Team. R: A language and environment for statistical computing (R Foundation for Statistical Computing, 2018).
Venables, W. N. & Ripley, B. D. Modern Applied Statistics with S 4th edn, (Springer, 2002).
Perez-Riverol, Y. et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019).
Wojtasz, L. et al. Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet. 5, e1000702 (2009).
Gaysinskaya, V. & Bortvin, A. Flow cytometry of murine spermatocytes. Curr. Protoc. Cytom. 72, 7 44.1–7.44.24 (2015).
Chong, L. et al. A human telomeric protein. Science 270, 1663–1667 (1995).
Tres, L. L. Extensive pairing of the XY bivalent in mouse spermatocytes as visualized by whole-mount electron microscopy. J. Cell Sci. 25, 1–15 (1977).
Bergs, J. W. et al. Differential expression and sex chromosome association of CHD3/4 and CHD5 during spermatogenesis. PLoS ONE 9, e98203 (2014).
Kourmouli, N. et al. Heterochromatin and tri-methylated lysine 20 of histone H4 in animals. J. Cell Sci. 117, 2491–2501 (2004).
Peters, A. H. et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323–337 (2001).
Turner, J. M., Burgoyne, P. S. & Singh, P. B. M31 and macroH2A1.2 colocalise at the pseudoautosomal region during mouse meiosis. J. Cell Sci. 114, 3367–3375 (2001).
Costa, Y. et al. Two novel proteins recruited by synaptonemal complex protein 1 (SYCP1) are at the centre of meiosis. J. Cell Sci. 118, 2755–2762 (2005).
Acknowledgements
We thank A. Tóth and B. de Massy for antibodies, mice, discussions and sharing of unpublished information; A. North and the Bio-Imaging Resource Center at Rockefeller University for assistance with SIM (supported by award number S10 RR031855 from the National Center For Research Resources); and R. Hendrickson, R. Soni and Z. Li (MSKCC Proteomics Core) for assistance with mass spectrometry. This work used the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov). MSKCC core facilities are supported by a Cancer Center Support Grant (P30 CA008748). L.A. was supported in part by a fellowship from the Lalor Foundation; M.B. was supported in part by a Rubicon fellowship from the Netherlands Organization for Scientific Research; and M.v.O. was supported in part by an NIH fellowship (F32 GM096692). This work was supported by NIGMS grants R35 GM118092 (S.K.) and R35 GM118175 (M.J.).
Author information
Authors and Affiliations
Contributions
L.A. designed and conducted all of the cytogenetic experiments presented and analysed the data; M.E.K. generated Ankrd31 mutant mice and anti-ANKRD31 antibodies; M.B. and M.E.K. provided Ankrd31 mutant mice and unpublished data; M.E.K. performed the immunoprecipitation for mass spectrometry and T.L. validated the ANKRD31-interacting proteins; K.B. and F.P. performed SSDS and analysed the data under the supervision of R.D.C.-O., with input from L.A. and S.K.; M.v.O. generated REC8 and REC114 antibodies; L.K. performed initial characterization and provided unpublished data on the PAR ultrastructure and cohesin enrichment; M.J. and S.K. designed and supervised the research, analysed the data and secured funding; L.A. and S.K. wrote the manuscript with input from M.J. All authors edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Francesca Cole Bergemann and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 PAR axis thickening and accumulation of RMMAI proteins.
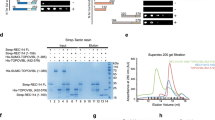
a, Axis thickening (SYCP3 and HORMAD2 staining) at the PAR (arrowheads) in a late-zygotene spermatocyte. HORMAD2 staining in the PAR at late zygonema mimics SYCP3 staining in all late-zygonema spermatocytes analysed (n > 20) in three mice. Scale bars, 2 μm. b, Image (adapted under a Creative Commons CC-BY licence from a previous study45) showing enrichment of HORMAD1 on the thick PAR axis of the Y chromosome. c, Colocalization of ANKRD31 and MEI4, REC114, IHO1 and MEI1. Representative zygotene spermatocytes are shown. Arrowheads indicate densely staining blobs. Areas indicated by dashed boxes are shown at higher magnification. The graphs show the total number of foci colocalized in leptotene or zygotene spermatocytes (mean ± s.d.). N.D., not determined: the low immunofluorescence signal for MEI1 did not allow us to quantify the colocalization with ANKRD31, although MEI1 showed clear colocalization with ANKRD31 in the blobs and at least some autosomal foci (insets). Scale bars, 2 μm. d, PARb FISH probe colocalizes with REC114 blobs. Two blobs are on the PAR, as judged by chromosome morphology and bright FISH with a PARb probe, and others highlight specific autosome ends. The colocalization between REC114 blobs and PARb FISH signals was observed in all spermatocytes analysed (n > 60), from pre-leptonema to early pachynema, in more than three mice. Scale bar, 2 μm. e, ANKRD31, REC114 and MEI1 immunostaining starts to appear in pre-leptonema. Seminiferous tubules were cultured with EdU to label replicating cells, and then chromosome spreads were stained for SYCP3 and either MEI1 with REC114 or ANKRD31 with PARb FISH. Colocalized foci appear in pre-leptonema (EdU-positive cells that are weakly SYCP3 positive), as previously shown for MEI4 and IHO117,22. Because we can already detect ANKRD31 accumulation at sites of PARb hybridization, we infer that the stronger sites of accumulation of MEI1 and REC114 also include PARs. PARb colocalized with ANKRD31 blobs (top) and MEI1 with REC114 (bottom) in all pre-leptotene spermatocytes analysed (n > 20) in one mouse. Scale bars, 2 μm. f, REC114 is not detected in the mo-2 regions in spermatogonia. Seminiferous tubules were cultured with EdU, and chromosome spreads were stained for DMRT1 (a marker of spermatogonia46) and REC114 plus mo-2 FISH. REC114 blobs colocalized with mo-2 FISH signals in the pre-leptotene spermatocyte (bottom) but were not apparent in the DMRT1-positive spermatogonium (top). Both cells shown were captured in a single microscopic field. Mo-2 FISH signals do not colocalize with the REC114 signal in all of the spermatogonia analysed (n > 20) in one mouse. Scale bar, 2 μm. g, Candidate ANKRD31-interacting proteins. To identify other PAR-associated proteins, ANKRD31 was immunoprecipitated from extracts made from whole testes of 12-day-old mice using two different polyclonal antibodies. This table shows a subset of proteins that were identified by mass spectrometry in immunoprecipitates from the testes of wild-type but not Ankrd31−/− mice, and not in immunoprecipitates using an irrelevant antibody (anti-cyclin B3). LFQ, label-free quantification. h, Enrichment of ZMYM3 (top) and PTIP (bottom) at the PAR. Sex chromosomes of representative early-pachytene spermatocytes are shown. ZMYM3 and PTIP were enriched in the PAR in all spermatocytes analysed (n > 20) in three mice. Scale bars, 2 μm. i, Yeast two-hybrid assays testing the interaction of full-length (FL) ANKRD31 fused to Gal4AD with either full-length PTIP or the C-terminal 191 amino acids of ZMYM3 fused to Gal4BD (full-length ZMYM3 autoactivates in this assay). Double dropout (DDO) medium selects for the presence of both the Gal4AD and the Gal4BD vector (positive control for growth); quadruple dropout (QDO) and QXA (QDO plus X-α-gal and aureobasidin A) media select for a productive two-hybrid interaction at lower and higher stringency, respectively. The image is representative of two experiments using the same yeast strains.
Extended Data Fig. 2 PAR ultrastructure.
a, Comparison of conventional microscopy and SIM, showing that the thickened PAR axis in conventional microscopy is resolved as separated axial cores (arrowheads). The thickening of the PAR axis in conventional microscopy and the splitting of the PAR axis in SIM was observed in more than 60 spermatocytes at late zygonema in at least three mice. Scale bars, 2 μm. b, Ultrastructure of the axis proteins SYCP2, SYCP3 and HORMAD2 in the PAR. SYCP2 (left) and HORMAD2 (right) staining mimic SYCP3 staining in late zygonema in all cells analysed (n > 30 in at least three mice by conventional microscopy; n = 5 in one mouse by SIM) (except that HORMAD2 appears rather depleted at the telomeres compared to SYCP3 and SYCP2). Scale bars, 1 μm. c–e, Distinguishing between splitting apart of sister chromatid axes and a crozier configuration in which a single conjoined axis for both sister chromatids is folded back on itself. A crozier (cartooned in c) was ruled out because the telomere-binding protein TRF147 decorates the tip of the PAR bubble (d) and the FISH signal for the PARb probe is arrayed relatively symmetrically on both axial cores (e), consistent with separated sister chromatid axes. Scale bars, 1 μm. The presence of TRF1 at the distal tip of the PAR was observed in all spermatocytes analysed (n > 20 by conventional microscopy; n = 3 by SIM) in one mouse. PARb FISH signals were relatively symmetrically arranged along the split PAR axes (n > 100 by conventional microscopy; n = 9 by SIM) in three mice. f, Schematic of the PAR ultrastructure and distribution of axis and RMMAI proteins at late zygonema. Tel., telomere. g, h, Paired PARs with elongated and split axes occur in late zygonema to early pachynema. Electron micrographs (adapted with permission from a previous study48) are shown in comparison with SIM immunofluorescence images of spermatocytes at early pachynema (g) or late zygonema (h; cyan arrowheads indicate examples of incomplete autosomal synapsis). Elongation and splitting of PAR axes occurs earlier than originally thought48 (Supplementary Discussion). Scale bars in SIM images, 1 μm (g); 2 μm (h). Extended and split PAR axes were observed by SIM (n > 30 spermatocytes) around the zygonema–pachynema transition in more than three mice. i, REC114 enrichment and axis splitting occurs in the absence of SPO11, thus neither is provoked by DSB formation. PAR axis splitting and extension of the RMMAI signal were observed by SIM in Spo11−/− mice in more than 20 late-zygotene-like spermatocytes in more than three mice. The differentiation of the PAR axis became hardly detectable at later stages in some pachytene-like spermatocytes as cells entered apoptosis. Scale bar, 1 μm.
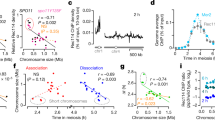
Extended Data Fig. 3 Time course of the spatial organization of the PAR loop–axis ensemble.
a, Time course of REC8 and ANKRD31 immunostaining along the PAR axis from pre-leptonema (preL, left) to mid-pachynema (right). A montage of representative SIM images is shown. Chromosomes a–e are presumptive X or Y, but could be the distal end of chromosome 9. Chromosomes at later stages were unambiguously identified by morphology. Chromosomes i–k show examples in which the initial pairing (probably synaptic) contact between X and Y chromosomes is centromere-proximal (that is, closer to the PAR boundary) (i), distal (closer to the telomere) (k), or interstitial (j). The preferential enrichment of REC8 at the border of the PAR split axes was observed in more than 30 zygotene spermatocytes by SIM in more than three mice. Scale bar, 1 μm. b, We collected three measurements of conventional immuno-FISH images from leptonema through to mid-pachynema: length of the REC114 signal along the PAR axis; maximum distance from the PARb FISH signal to the distal end of the SYCP3-defined axis; and axis-orthogonal extension of the FISH signal for the PARb probe (a proxy for loop sizes). Data were collected from three males. Insets show examples of each type of measurement at each stage. Horizontal black lines indicate means. Means of each measurement for each mouse at each stage are given below, along with the means across all three mice. Means are rounded to two significant figures; the grand means were calculated using unrounded values from individual mice. The number of cells of each stage from each mouse is given. Modest variability in the apparent dimensions of the Y chromosome PAR between different mice may be attributable to variation in the copy number of mo-2 and other repeats because of unequal exchange during meiosis. Nonetheless, highly similar changes in spatial organization over time in prophase were observed in all mice examined—namely, progressive elongation then shortening of axes and concomitant lengthening of loops. Scale bar, 1 μm. In brief, a and b show the following. At pre-leptonema, ANKRD31 blobs had a closely juxtaposed focus of the meiotic cohesin subunit REC8 (chromosome a). In leptonema and early zygonema, ANKRD31 and REC114 signals stretched along the presumptive PAR axes, with REC8 restricted to the borders (a, chromosomes b–e). The SYCP3-defined axis was already long as soon as it was detectable (0.73 μm) and the PARb FISH signal was compact (0.52 μm) (b, i). At late zygonema, the PAR axis had lengthened still further (1.0 μm), whereas the PARb signal remained compact (b, ii). The PAR split into separate axes during this stage, each with abundant RMMAI (a, chromosomes f–h). The split was a REC8-poor zone bounded by REC8 foci (a, chromosomes f–h; Extended Data Fig. 2f). After synapsis, the axes shortened and chromatin loops decompacted, with concomitant dissociation of RMMAI proteins. As cells transitioned into early pachynema and the X and Y chromosome PARs synapsed (a, chromosomes i–m), the PAR axes began to shorten slightly (0.85 μm) and the PARb signal expanded (0.85 μm) (b, iii). Meanwhile, the elongated ANKRD31 signals progressively decreased in intensity, collapsed along with the shortening axes and separated from the axis while remaining nearby (a, chromosomes l–m). By mid-pachynema, the PAR axes collapsed still further, to about half their zygotene length (0.50 μm) and the PARb chromatin expanded to more than twice the zygotene measurement (1.3 μm). ANKRD31 and REC114 enrichment largely disappeared, leaving behind a bright bolus of REC8 on the short remaining axis (a, chromosomes n–o; b, iv). c, Non-homologous synapsis appears sufficient to trigger collapse of the PAR loop–axis structure. We measured the length of the REC114 signal along the PAR axis and the extension of mo-2 chromatin orthogonal to the axis in Spo11−/− spermatocytes in which the X chromosome PAR had non-homologously synapsed with an autosome while the Y chromosome PAR remained unsynapsed. Within any given cell, the unsynapsed Y chromosome PAR maintained the characteristic late-zygotene configuration (long axis, short loops) whereas the synapsed X chromosome PAR adopted the configuration characteristic of pachynema (short axis, long loops). Error bars are mean ± s.d. Scale bar, 2 μm.
Extended Data Fig. 4 RMMAI enrichment at mo-2 minisatellite arrays in the PAR and on specific autosomes.
a, Top, self-alignment of the PARb FISH probe (reproduced from Fig. 2a). The circled block is a 20-kb mo-2 cluster. Bottom, schematic depicting the last 1.4 Mb of the non-centromeric ends of the indicated chromosomes, showing the positions of mo-2 repeats (green) adjacent to assembly gaps (mm10); mo-2 repeats were identified by BLAST search using the mo-2 consensus sequence. Mo-2 repeats also appear at the distal end of chromosome 4 in the Celera assembly (Mm_Celera, 2009/03/04). PARb and PARd BAC clones are indicated. b, Confirmation that autosomal mo-2 FISH signals match the chromosomal locations indicated by mm10 or Celera genome assemblies. FISH was performed using an oligonucleotide probe containing the mo-2 consensus sequence in combination with BAC probes for adjacent segments of chromosomes 13, 9 and 4, as indicated. Magenta arrows point to concordant FISH signals. The chromosome 9 BAC probe also hybridizes to the PAR. The colocalization of mo-2 and the three autosomal FISH signals was observed in two mice (n > 20 spermatocytes). Scale bars, 2 μm. c, Comparison of mo-2 FISH with REC114 localization relative to the PAR boundary (PARb FISH probe) and the distal PAR (PARd probe). In mid-zygonema, the mo-2 FISH signal colocalizes well with REC114 staining in between the PARb and PARd FISH signals. In late zygonema, mo-2 and REC114 are similar to one another and are elongated along the thickened SYCP3 staining of the PAR axis. From early to mid-pachynema, REC114 progressively disappears, whereas the mo-2 FISH signal becomes largely extended away from the PAR axes. The relative positions of the PARb and PARd probes reinforce the conclusion that the PAR does not adopt a crozier configuration. The different positioning of PARb and PARd FISH signals compared to mo-2 or REC114 signals was observed in more than 30 spermatocytes in at least three mice. Scale bar, 1 μm. d, Illustration of the compact organization of the PAR chromatin (mo-2 FISH signal) compared to a whole Y chromosome paint probe. The costaining of mo-2 and the full chromosome Y probe was evaluated in one mouse (n > 20 spermatocytes). Scale bar, 2 μm. e, Lower mo-2 copy number in the M. musculus molossinus subspecies correlates with lower REC114 staining in mo-2 regions. The left panels compare MSM and B6 mice for colocalization between REC114 immunostaining and mo-2 FISH in leptotene spermatocytes. The REC114 and SYCP3 channels are shown at equivalent exposure for the two strains, whereas a longer exposure is shown for the mo-2 FISH signal in the MSM spermatocyte. The mo-2-associated REC114 blobs are much brighter relative to the smaller dispersed REC114 foci in the B6 spermatocyte than in the MSM spermatocyte. The right panel shows representative pachytene spermatocytes to confirm the locations of mo-2 clusters at autosome ends and at the PAR in the MSM background. The lower intensity of REC114 blobs in MSM compared to B6 spermatocytes was observed in more than 30 spermatocytes in three different pairs of mice. Scale bars, 2 μm. f, PAR enrichment for ANKRD31 and RPA2 correlates with mo-2 copy number. Top, late-zygotene spermatocytes from MSM × B6 F1 hybrids. Scale bar, 1 μm. Bottom, PAR-associated signals on B6-derived (YB6) and MSM-derived (XMSM) chromosomes from the indicated number of spermatocytes. Red lines indicate mean ± s.d. Differences between the PAR intensities of X and Y chromosomes are significant both for proteins and for mo-2 FISH in both F1 hybrids (P < 10−13, paired t-test; for exact P values, see the associated Source Data). g, Representative micrographs of late-zygotene spermatocytes from reciprocal F1 hybrid males from crosses of B6 (high mo-2 copy number) and MSM (low mo-2 copy number) parents. Scale bars, 1 μm. h, Frequency of paired X and Y chromosomes at late zygonema and mid-pachynema analysed in three MSM and three B6 males. Differences between strains were not statistically significant at either stage (P = 0.241 for late zygonema and P = 0.136 for mid-pachynema; two-sided Student’s t-test). X and Y are late-pairing chromosomes in the MSM background—as in the B6 background. The similar pairing kinetics indicates that the lower intensity of RMMAI staining on the MSM PAR is not attributable to earlier PAR pairing and synapsis in this strain. The number of spermatocytes analysed is indicated.
Extended Data Fig. 5 Mo-2 regions accumulate heterochromatin factors.
a, Costaining of ANKRD31 or mo-2 with the indicated proteins and histone marks known to localize at the pericentromeric heterochromatin (mouse major satellite), in zygotene spermatocytes (left) and pre-leptotene spermatocytes (right). Each of the heterochromatin factors shows a locally enriched signal coincident with mo-2 regions (arrowheads), in addition to broader staining of other sub-nuclear regions. The CHD3/4 antibody recognizes both proteins49. The colocalization of ANKRD31 blobs with heterochromatin blobs was observed in all zygotene spermatocytes analysed (n > 20) in at least three mice for each antibody (left) and in one mouse for pre-leptotene spermatocytes (n > 10) for each antibody (right). Scale bars, 2 μm. b, CHD3/4, ATRX, HP1β, H4K20me3, H3K9me3 and macroH2A1.2 are not detectably enriched at mo-2 regions in spermatogonia (small, DMRT1-positive cells). These factors may be present at mo-2 regions in these cells, but do not appear to accumulate to elevated levels. The absence of colocalization between mo-2 FISH signals and heterochromatin factors was noted in all spermatogonia analysed (n > 30) from one mouse. Scale bars, 2 μm. c, Heterochromatin factors can be detected in the PAR up to late pachynema. Each of the assayed proteins and histone marks showed staining on the autosomal and X-specific pericentromeric heterochromatin, the sex body and euchromatin—albeit with variations between sites in the timing and level of accumulation. Notably, however, they also showed enriched staining at all mo-2 regions up to early or mid-pachynema, as shown for H4K20me3 (top). By mid-to-late pachynema, as shown here for H3K9me3, the signal persisted in the PAR but was usually barely detectable at mo-2 regions of chromosome 9 or chromosome 13. This observation indicates that—at least for the PAR—the heterochromatin factors can continue to be enriched on mo-2 chromatin after RMMAI proteins have dissociated. These results substantially extend previous observations about CHD3/4 colocalizing with PAR FISH signals; H4K20me3 being localized in the PAR and at the ends of other chromosomes; and detection of H3K9me3, HP1β and macroH2A1.2 in the PAR in late pachynema49,50,51,52. The colocalization between major satellite (maj sat) and H4K20me3 and H3K9me3 was observed in all spermatocytes analysed (n > 20) in one mouse. The colocalization between H4K20me3 and mo-2 FISH signals was observed in all spermatocytes analysed (n > 60) from the pre-leptotene to mid-pachytene stage in more than three mice. Scale bars, 2 μm. d, Enrichment of the heterochromatin factors is independent of SPO11. Representative images of Y chromosomes from a Spo11–/– mouse are shown. The colocalization between PAR mo-2 FISH signals and heterochromatin factors was observed in all Spo11−/− spermatocytes analysed (n > 30) in more than three mice for CHD3/4 and at least one mouse each for ATRX, HP1β, HP1γ, macroH2A1.2, H3K9me3 and H4K20me3. Scale bar, 1 μm.
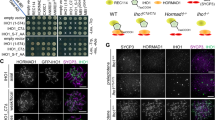
Extended Data Fig. 6 Genetic requirements for RMMAI assembly on chromosomes and for PAR loop–axis organization.
a, Representative micrographs of ANKRD31, MEI4, IHO1 and MEI1 staining in wild-type spermatocytes and the indicated mutants (quantification is in Fig. 3a). Scale bars, 2 μm. b, Measurements of PAR loop–axis organization, as in Fig. 3b, in two additional males. The data from mouse 1 are reproduced from Fig. 3b to facilitate comparison. Means of each measurement for each mouse at each stage are given below, along with the means across all three mice. Means are rounded to two significant figures; the grand means were calculated using unrounded values from individual mice. The number of cells of each stage from each mouse is given. c, REC8 is dispensable for the splitting apart of PAR sister chromatid axes, but is required to maintain the connection between sisters at the distal tip of the chromosome. A representative SIM image is shown of a Y chromosome from a late-zygotene Rec8−/− spermatocyte. The SYCP3-labelled axes adopt an open-fork configuration. The distal FISH probe (PARd) shows that there are clearly disjoined sisters, whereas the PAR boundary (PARb) shows only a single compact signal comparable to the wild type. The disposition of the probes and SYCP3 further rules out the crozier configuration as an explanation for split PAR axes. The structure of the Y or X chromosome PAR was resolved by SIM as ‘fork-shaped’ in all spermatocytes analysed (n > 20) from three mice. Scale bar, 1 μm. d, Quantification of REC114 and MEI4 foci in two additional pairs of wild-type and Ankrd31−/− mice. Horizontal lines indicate means. Fewer foci were observed in the Ankrd31−/− mutant (two-sided Student’s t-tests for each comparison of mutant to wild type: P = 5.6 × 10−6 (second set, REC114); P = 1.1 × 10−5 (second set, MEI4); P = 2.1 × 10−6 (third set, REC114); P = 0.017 (third set, MEI4)). e, Reduced REC114 staining intensity of axis-associated foci in Ankrd31−/− mutants. To rigorously control for slide-to-slide and within-slide variation in immunostaining, we mixed together wild-type and Ankrd31−/− testis cell suspensions before preparing chromosome spreads. A representative image is shown of a region from a single microscopic field containing two wild-type zygotene spermatocytes (left) and two Ankrd31−/− spermatocytes of the equivalent stage (right). Note the diminished intensity of REC114 foci in the Ankrd31−/− spermatocytes. Scale bar, 2 μm. REC114 (non-blob) foci showed a lower fluorescence intensity in Ankrd31–/– mutant compared to wild-type spermatocytes in all pairs of spermatocytes captured in the same imaging field (n = 8 pairs), from one pair of mice. f, PAR enrichment of heterochromatin-associated factors is independent of ANKRD31. Representative images of the Y chromosome at late zygonema or early pachynema, showing colocalization between the decompacted mo-2 chromatin and the indicated proteins. Both the FISH and the immunofluorescence signals are localized mostly off the axis (compare with the same signals in the absence of SPO11; Extended Data Fig. 5d). The mo-2 FISH signal colocalized off the axis with the heterochromatin factors in Ankrd31−/− mice in all spermatocytes analysed (n > 30) in more than three mice for CHD3/4 and at least one mouse for ATRX, HP1β, HP1γ, macroH2A1.2, H3K9me3 and H4K20me3. Scale bar, 1 μm.
Extended Data Fig. 7 PAR-associated RPA2 foci.
a, Loop–axis organization of the mo-2 region of chromosome 9 in late zygonema. Compare with the PAR (Fig. 3b). Error bars are mean ± s.d. Scale bars, 1 μm. b, Low mo-2 copy number correlates with less loop–axis reorganization (SIM images of late-zygotene F1 hybrid spermatocytes). The differentiation of the B6 PAR was observed in both hybrids (B6 × MSM and MSM × B6) in three and four spermatocytes, respectively, by SIM (one mouse for each), and in more than 20 spermatocytes by conventional microscopy (two mice of each genotype). Scale bars, 1 μm. c–e, Immuno-FISH for RPA2 and mo-2 was used to detect DSBs cytologically in wild-type mice and the indicated mutants. To analyse Rec8 and Hormad1 mutations, we compared to mutants lacking SYCE1 (a synaptonemal complex central element component53) because Syce1–/– mutants show similar meiotic progression defects without defective RMMAI recruitment. c, Representative images. Scale bars, 2 μm (main image); 1 μm (inset). d, Global counts of RPA2 foci for zygotene (zyg) or zygotene-like cells and for pachytene (pach) or pachytene-like cells. e, Fraction of mo-2 regions, for each cell, that had a colocalized RPA2 focus. Red lines indicate mean ± s.d. Statistical significance is indicated in d, e for comparisons (two-sided Student’s t-tests) of wild type to Ankrd31−/− or of Syce1−/− to either Rec8−/− or Hormad1−/− for matched stages; for exact P values, see the associated Source Data. The number of discretely scorable mo-2 regions in e varied from cell to cell depending on pairing status. f, Frequent formation of DSBs at mo-2 regions in the PAR and on autosomes does not require HORMAD1. Left, micrograph showing two adjacent spermatocytes (boundary indicated by dashed line). Scale bar, 2 μm. Right, insets show higher-magnification views of the numbered mo-2 regions, all of which are associated with RPA2 immunostaining of varying intensity. This image illustrates the preferential formation of RPA2 foci in mo-2 regions in a Hormad1−/− mouse; quantification is in e. g, Autosomal mo-2 regions often form DSBs late. Immuno-FISH for RPA2, mo-2 and PARb was used to detect DSBs cytologically in wild-type mice from leptonema to mid-pachynema, and to distinguish the X or Y chromosome PAR from chromosomes 9 and 13. Chromosome 4 was not assayed because the mo-2 FISH signal was often barely detectable. Top, global number of RPA2 foci per cell. Black lines indicate mean ± s.d. Bottom, percentage of spermatocytes with an RPA2 focus overlapping the PAR (X, Y or both) or overlapping chromosome 9 or 13. A representative image of an early-pachytene spermatocyte is shown. As previously shown for the PAR2, autosomal mo-2 regions continue to accumulate RPA2 foci beyond the time point at which global RPA2 foci have largely or completely ceased accumulating. Scale bar, 2 μm. h, X–Y chromosome pairing status, quantified by immuno-FISH for SYCP3 and the PARd probe. i, Montage of SIM images from a B6 male, showing that multiple, distinct RPA2 foci can be detected from late zygonema to mid-pachynema. This suggests that multiple PAR DSBs can be formed during one meiosis2. The presence of multiple RPA2 foci in the PAR was observed by SIM in more than 20 spermatocytes from late zygonema to mid-pachynema in one mouse. Scale bar, 1 μm. j, Percentage of spermatocytes at the zygotene–pachytene transition with no (0), one, two or three distinguishable RPA2 foci on the unsynapsed Y chromosome PAR of MSM and B6 mice. The difference between the strains is statistically significant (negative binomial regression, P = 7.2 × 10−5). The number of spermatocytes analysed is indicated. A representative image is shown for each genotype, with one RPA2 focus on the MSM PAR and two apparent sites of RPA2 accumulation on the B6 PAR. The detection of multiple foci is consistent with reported double crossovers6. Scale bar, 1 μm.
Extended Data Fig. 8 DSB maps on the PAR and autosomal mo-2 regions.
a, SSDS sequence coverage (data from previous reports7,20) is shown for the X chromosome PAR (shown in a different form previously20), the Y chromosome PAR and the mo-2-adjacent regions of chromosomes 9 and 13. The dashed segments indicate gaps in the mm10 genome assembly. We did not assess chromosome 4 because available assemblies are too incomplete. b, Regions adjacent to the mo-2 region on chromosome 9 show an SSDS signal that is reproducibly increased relative to the chromosome 9 average in wild-type testis samples but not in maps from Ankrd31−/− testes or wild-type ovaries. Two of the SSDS browser tracks are reproduced from a. The bar chart shows enrichment values from individual SSDS maps (T1–T9 are maps from wild-type testes; O1 and O2 are from wild-type ovaries31). Enrichment values are defined as coverage across the indicated coordinates relative to the mean coverage for chromosome 9 (see Methods for details). Ovary sample O1 and the Ankrd31−/− adult sample are known to have poorer signal-to-noise ratios than the other samples20,31. For all SSDS coverage tracks, reads that map to multiple locations are included after random assignment to one of their mapped positions. However, the same conclusions are reached about ANKRD31 dependence and PRDM9 independence of the signal on chromosomes 9 and 13 if only uniquely mapped reads are used. c, Oocytes incur substantially fewer DSBs than spermatocytes near the mo-2 region on chromosome 9. The SSDS signal is from a previous study31 (samples T1 and O2). The X chromosome PAR is shown for comparison (previously shown to be essentially devoid of DSBs in ovary samples31). See b for quantification.
Extended Data Fig. 9 RMMAI accumulation and low frequency of DSB formation at mo-2 regions in oocytes.
a, Examples of zygotene oocytes showing the colocalization between blobs of IHO1 and REC114, MEI4 and MEI1, or ANKRD31 and mo-2 FISH signal (arrowheads). RMMAI blobs colocalized with mo-2 FISH signals in all zygotene oocytes analysed (n > 30) from at least three mice. Scale bars, 2 μm. b, PAR ultrastructure in oocytes, quantified as in Extended Data Fig. 3b. Late-zygotene cells with PAR synapsis are compiled separately from other zygotene cells. Error bars are mean ± s.d. Scale bar, 1 μm. c, Examples of zygotene oocytes showing colocalization of ANKRD31 blobs with enrichment for heterochromatin factors. ANKRD31 colocalized with blobs of heterochromatin factors in all zygotene oocytes analysed (n > 20) from one mouse. Scale bars, 2 μm. d, Representative SIM image of a wild-type late-zygotene oocyte showing neither detectable splitting of the PAR axis nor REC8 enrichment. The absence of spermatocyte-like differentiation of the PAR axis was observed (n > 30 zygotene oocytes) in more than three mice. A modest degree of differentiation was observed in a minority of oocytes (5/45) analysed by SIM, but this did not resemble the typical PAR axis splitting found in spermatocytes. Scale bar, 2 μm. e, Prolonged asynapsis does not allow axis splitting to occur in oocytes. Because synapsis appears to be sufficient to trigger the collapse of the PAR ultrastructure in spermatocytes (Extended Data Fig. 3b), we asked whether preventing synapsis (that is, in a Syce1−/− mutant) could reveal a tendency towards axis splitting in oocytes. Whereas axis splitting was clearly observed by SIM in Syce1−/− mutant spermatocytes, PAR axes were not detectably split in oocytes. Axis splitting of chromosome 9 was observed by SIM in multiple (n > 20) Syce1−/− spermatocytes from three different mice. The centromere-distal axes were also occasionally seen to be split in chromosomes 13 or 4, but we did not quantify this for these chromosomes. In males, the differentiation of the axes of the PAR or chromosome 9 becomes hardly detectable at later stages in some pachytene-like spermatocytes as cells enter apoptosis, similar to Spo11−/− or Hormad1−/− mice. However, in Syce1−/− oocytes, no obvious axis differentiation or splitting was observed by conventional microscopy or by SIM in multiple oocytes (n > 30) from three different mice—similar to what was observed in wild-type oocytes. Scale bars, 2 μm (main image); 1 μm (insets). f, h, Delaying synapsis promotes the formation of DSBs in the PAR in oocytes. Top, representative micrographs of pachytene XY (f) and Syce1−/− XX (h) oocytes. Middle, RPA2 fluorescence intensity at the border of mo-2 FISH signals from the PAR, chromosome 9 and chromosome 13. Bottom, percentage of oocytes with an RPA2 focus colocalizing with mo-2 regions on the PAR, chromosome 9 and chromosome 13. Graphs show data only for pachytene oocytes in which PARs are synapsed (two mice of each genotype). Red lines indicate mean ± s.d. Scale bars, 2 μm. g, Percentage of pachytene oocytes with one or more RPA2 foci colocalizing with the mo-2 FISH signal from the PAR, chromosome 9 and chromosome 13 in XY pachytene oocytes that had unsynapsed X and Y chromosomes. Scale bars, 2 μm (main image); 1 μm (inset).
Extended Data Fig. 10 Summary of PAR ultrastructure and molecular determinants of axis remodelling and DSB formation.
Schematic of the meiotic Y chromosome loop–axis structure before the pairing or synapsis of X and Y chromosomes at the transition between zygonema and pachynema. The chromosome axis comprises the meiosis-specific axial proteins SYCP2, SYCP3, HORMAD1 and HORMAD2; cohesin subunits (only REC8 is represented); and RMMAI proteins (REC114, MEI4, MEI1, ANKRD31 and IHO1). On the non-PAR portion of the Y chromosome axis (left), RMMAI protein loading and DSB formation are partly dependent on HORMAD1 and ANKRD31, and strictly dependent on MEI4, REC11419, IHO121 and, presumably, MEI118. The DNA is organized into large loops, with a low number of axis-associated RMMAI foci. By contrast, in the PAR (right), the hyperaccumulation of RMMAI proteins at mo-2 minisatellites (possibly spreading into the adjacent chromatin) promotes the elongation and subsequent splitting of the PAR sister chromatid axes. Short mo-2-containing chromatin loops stretch along this extended PAR axis, which increases the physical distance between the PAR boundary and the distal PAR sequences, including the telomere. The degree of RMMAI protein loading, PAR axis differentiation and DSB formation are proportional to the mo-2 FISH signal (which we interpret as reflecting mo-2 copy number), and depend on MEI4, ANKRD31 and, presumably, REC114.
Supplementary information
Supplementary Information
This file contains the Supplementary Discussion and Supplementary References.
Rights and permissions
About this article
Cite this article
Acquaviva, L., Boekhout, M., Karasu, M.E. et al. Ensuring meiotic DNA break formation in the mouse pseudoautosomal region. Nature 582, 426–431 (2020). https://doi.org/10.1038/s41586-020-2327-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2327-4
This article is cited by
-
Divergence and conservation of the meiotic recombination machinery
Nature Reviews Genetics (2024)
-
Seeding the meiotic DNA break machinery and initiating recombination on chromosome axes
Nature Communications (2024)
-
Bi-allelic MEI1 variants cause meiosis arrest and non-obstructive azoospermia
Journal of Human Genetics (2023)
-
The proper interplay between the expression of Spo11 splice isoforms and the structure of the pseudoautosomal region promotes XY chromosomes recombination
Cellular and Molecular Life Sciences (2023)
-
The RNA-binding protein FUS/TLS interacts with SPO11 and PRDM9 and localize at meiotic recombination hotspots
Cellular and Molecular Life Sciences (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.