Abstract
Mitochondria take up Ca2+ through the mitochondrial calcium uniporter complex to regulate energy production, cytosolic Ca2+ signalling and cell death1,2. In mammals, the uniporter complex (uniplex) contains four core components: the pore-forming MCU protein, the gatekeepers MICU1 and MICU2, and an auxiliary subunit, EMRE, essential for Ca2+ transport3,4,5,6,7,8. To prevent detrimental Ca2+ overload, the activity of MCU must be tightly regulated by MICUs, which sense changes in cytosolic Ca2+ concentrations to switch MCU on and off9,10. Here we report cryo-electron microscopic structures of the human mitochondrial calcium uniporter holocomplex in inhibited and Ca2+-activated states. These structures define the architecture of this multicomponent Ca2+-uptake machinery and reveal the gating mechanism by which MICUs control uniporter activity. Our work provides a framework for understanding regulated Ca2+ uptake in mitochondria, and could suggest ways of modulating uniporter activity to treat diseases related to mitochondrial Ca2+ overload.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The three-dimensional cryo-EM density maps are deposited into the Electron Microscopy Data Bank (https://www.ebi.ac.uk/pdbe/emdb/) under accession numbers EMD-21642 and EMD-21643. The coordinates are deposited in the Protein Data Bank (https://www.rcsb.org) with accession numbers 6WDN and 6WDO.
References
Rizzuto, R., De Stefani, D., Raffaello, A. & Mammucari, C. Mitochondria as sensors and regulators of calcium signalling. Nat. Rev. Mol. Cell Biol. 13, 566–578 (2012).
Giorgi, C., Marchi, S. & Pinton, P. The machineries, regulation and cellular functions of mitochondrial calcium. Nat. Rev. Mol. Cell Biol. 19, 713–730 (2018).
Kirichok, Y., Krapivinsky, G. & Clapham, D. E. The mitochondrial calcium uniporter is a highly selective ion channel. Nature 427, 360–364 (2004).
De Stefani, D., Raffaello, A., Teardo, E., Szabò, I. & Rizzuto, R. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature 476, 336–340 (2011).
Baughman, J. M. et al. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature 476, 341–345 (2011).
Perocchi, F. et al. MICU1 encodes a mitochondrial EF hand protein required for Ca2+ uptake. Nature 467, 291–296 (2010).
Plovanich, M. et al. MICU2, a paralog of MICU1, resides within the mitochondrial uniporter complex to regulate calcium handling. PLoS ONE 8, e55785 (2013).
Sancak, Y. et al. EMRE is an essential component of the mitochondrial calcium uniporter complex. Science 342, 1379–1382 (2013).
Kamer, K. J. & Mootha, V. K. The molecular era of the mitochondrial calcium uniporter. Nat. Rev. Mol. Cell Biol. 16, 545–553 (2015).
De Stefani, D., Rizzuto, R. & Pozzan, T. Enjoy the trip: calcium in mitochondria back and forth. Annu. Rev. Biochem. 85, 161–192 (2016).
Mallilankaraman, K. et al. MICU1 is an essential gatekeeper for MCU-mediated mitochondrial Ca2+ uptake that regulates cell survival. Cell 151, 630–644 (2012).
Csordás, G. et al. MICU1 controls both the threshold and cooperative activation of the mitochondrial Ca2+ uniporter. Cell Metab. 17, 976–987 (2013).
Patron, M. et al. MICU1 and MICU2 finely tune the mitochondrial Ca2+ uniporter by exerting opposite effects on MCU activity. Mol. Cell 53, 726–737 (2014).
Petrungaro, C. et al. The Ca2+-dependent release of the Mia40-induced MICU1-MICU2 dimer from MCU regulates mitochondrial Ca2+ uptake. Cell Metab. 22, 721–733 (2015).
Peng, T. I. & Jou, M. J. Oxidative stress caused by mitochondrial calcium overload. Ann. NY Acad. Sci. 1201, 183–188 (2010).
Logan, C. V. et al. Loss-of-function mutations in MICU1 cause a brain and muscle disorder linked to primary alterations in mitochondrial calcium signaling. Nat. Genet. 46, 188–193 (2014).
Musa, S. et al. A middle eastern founder mutation expands the genotypic and phenotypic spectrum of mitochondrial MICU1 deficiency: a report of 13 patients. JIMD Rep. 43, 79–83 (2018).
Tsai, M. F. et al. Dual functions of a small regulatory subunit in the mitochondrial calcium uniporter complex. eLife 5, e15545 (2016).
Kamer, K. J., Grabarek, Z. & Mootha, V. K. High-affinity cooperative Ca2+ binding by MICU1-MICU2 serves as an on-off switch for the uniporter. EMBO Rep. 18, 1397–1411 (2017).
Payne, R., Hoff, H., Roskowski, A. & Foskett, J. K. MICU2 restricts spatial crosstalk between InsP3R and MCU channels by regulating threshold and gain of MICU1-mediated inhibition and activation of MCU. Cell Rep. 21, 3141–3154 (2017).
Paillard, M. et al. MICU1 interacts with the D-ring of the MCU pore to control its Ca2+ flux and sensitivity to Ru360. Mol. Cell 72, 778–785 (2018).
Phillips, C. B., Tsai, C. W. & Tsai, M. F. The conserved aspartate ring of MCU mediates MICU1 binding and regulation in the mitochondrial calcium uniporter complex. eLife 8, e41112 (2019).
Fan, C. et al. X-ray and cryo-EM structures of the mitochondrial calcium uniporter. Nature 559, 575–579 (2018).
Baradaran, R., Wang, C., Siliciano, A. F. & Long, S. B. Cryo-EM structures of fungal and metazoan mitochondrial calcium uniporters. Nature 559, 580–584 (2018).
Nguyen, N. X. et al. Cryo-EM structure of a fungal mitochondrial calcium uniporter. Nature 559, 570–574 (2018).
Yoo, J. et al. Cryo-EM structure of a mitochondrial calcium uniporter. Science 361, 506–511 (2018).
Wang, Y. et al. Structural mechanism of EMRE-dependent gating of the human mitochondrial calcium uniporter. Cell 177, 1252–1261 (2019).
Wang, L. et al. Structural and mechanistic insights into MICU1 regulation of mitochondrial calcium uptake. EMBO J. 33, 594–604 (2014).
Xing, Y. et al. Dimerization of MICU proteins controls Ca2+ influx through the mitochondrial Ca2+ uniporter. Cell Rep. 26, 1203–1212 (2019).
Kamer, K. J., Jiang, W., Kaushik, V. K., Mootha, V. K. & Grabarek, Z. Crystal structure of MICU2 and comparison with MICU1 reveal insights into the uniporter gating mechanism. Proc. Natl Acad. Sci. USA 116, 3546–3555 (2019).
Wu, W. et al. The crystal structure of MICU2 provides insight into Ca2+ binding and MICU1-MICU2 heterodimer formation. EMBO Rep. 20, e47488 (2019).
Kamer, K. J. & Mootha, V. K. MICU1 and MICU2 play nonredundant roles in the regulation of the mitochondrial calcium uniporter. EMBO Rep. 15, 299–307 (2014).
Davies, K. M., Anselmi, C., Wittig, I., Faraldo-Gómez, J. D. & Kühlbrandt, W. Structure of the yeast F1Fo-ATP synthase dimer and its role in shaping the mitochondrial cristae. Proc. Natl Acad. Sci. USA 109, 13602–13607 (2012).
De La Fuente, S. et al. Strategic positioning and biased activity of the mitochondrial calcium uniporter in cardiac muscle. J. Biol. Chem. 291, 23343–23362 (2016).
Singh, A. K., McGoldrick, L. L., Twomey, E. C. & Sobolevsky, A. I. Mechanism of calmodulin inactivation of the calcium-selective TRP channel TRPV6. Sci. Adv. 4, eaau6088 (2018).
Dang, S. et al. Structural insight into TRPV5 channel function and modulation. Proc. Natl Acad. Sci. USA 116, 8869–8878 (2019).
Banerjee, A., Lee, A., Campbell, E. & Mackinnon, R. Structure of a pore-blocking toxin in complex with a eukaryotic voltage-dependent K+ channel. eLife 2, e00594 (2013).
Park, C. S. & Miller, C. Interaction of charybdotoxin with permeant ions inside the pore of a K+ channel. Neuron 9, 307–313 (1992).
Patron, M., Granatiero, V., Espino, J., Rizzuto, R. & De Stefani, D. MICU3 is a tissue-specific enhancer of mitochondrial calcium uptake. Cell Death Differ. 26, 179–195 (2019).
Rizzuto, R. et al. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science 280, 1763–1766 (1998).
Santulli, G., Xie, W., Reiken, S. R. & Marks, A. R. Mitochondrial calcium overload is a key determinant in heart failure. Proc. Natl Acad. Sci. USA 112, 11389–11394 (2015).
Starkov, A. A., Chinopoulos, C. & Fiskum, G. Mitochondrial calcium and oxidative stress as mediators of ischemic brain injury. Cell Calcium 36, 257–264 (2004).
Goehring, A. et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat. Protoc. 9, 2574–2585 (2014).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Grant, T., Rohou, A. & Grigorieff, N. cisTEM, user friendly software for single-particle image processing. eLife 7, e35383 (2018).
Scheres, S. H. W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Ohlendieck, K., Riesinger, I., Adams, V., Krause, J. & Brdiczka, D. Enrichment and biochemical characterization of boundary membrane contact sites from rat-liver mitochondria. Biochim. Biophys. Acta 860, 672–689 (1986).
Acknowledgements
We thank L. Montabana and D.-H. Chen at Stanford–SLAC cryo-EM facilities; C. Xu, K. Lee and K. Song at the University of Massachusetts cryo-EM facility for help with collecting EM data; A. Van Keuren for technical assistance; and S. R. Levinson for assistance in establishing the submitochondrial fractionation assay. This work was made possible by support from Stanford University and the Harold and Leila Y. Mathers Charitable Foundation to L.F.; an American Heart Association postdoctoral fellowship to M.F.; and a Dean’s fellowship to J.Z. M.-F.T., C.-W.T. and M.R. are supported by the National Institutes of Health (NIH) grant R01-GM129345.
Author information
Authors and Affiliations
Contributions
M.F. and J.Z. carried out biochemical experiments and cryo-EM studies. C.-W.T. carried out functional studies. B.J.O. contributed to EM studies. M.R. contributed to functional studies. Y.X. contributed to biochemical and functional characterizations. M.L. advised the EM studies. M.-F.T. oversaw the functional studies. L.F. directed the overall project. M.F., M.-F.T. and L.F. wrote the manuscript with input and support from J.Z., B.J.O., C.-W.T. and M.L.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Murali Prakriya, Alexander I. Sobolevsky and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Biochemical characterization of the purified human uniplex and validation of the interfaces of the low-Ca2+ uniplex.
a, Size-exclusion chromatography profile of the purified human uniplex. b, SDS–PAGE analysis of the purified human uniplex. The disulfide-linked MICU1–MICU2 heterodimer is labelled with an asterisk. Data in a, b are representative of five independent experiments with similar results. c, Effects of MCU–MICU1 interfacial mutations on complex stability. FLAG-tagged MICU1 was immobilized to pull down 1D4-tagged wild-type (WT) MCU co-expressed in MCU/EMRE/MICU1-knockout cells. IP, immunoprecipitate (eluted protein); WCL, whole-cell lysate. KA, RA and YA stand for K126A, R129A and Y114A mutations, respectively. In a separate control coimmunoprecipitation experiment, Letm1–FLAG was expressed alone in MCU/EMRE/MICU1-knockout cells, which were solubilized and incubated with FLAG beads. The eluent was then analysed. Letm1 in WCL and immunoprecipitate was detected using anti-FLAG and anti-Letm1 antibodies, respectively. Cytochrome c oxidase subunit 2 (COX2) serves as a control, showing that MICU1 does not interact nonspecifically with other mitochondrial inner-membrane proteins. MICU2 signals were obtained by targeting native proteins. An EMRE blot was not performed, as the EMRE gene was deleted in these cells. MICU1 mutants were properly folded: they still formed a complex with MICU2. d, Functional roles of MICU1’s C-terminal helix. In coimmunoprecipitation, wild-type MCU, wild-type MICU2 and FLAG-tagged MICU1 constructs were expressed in MCU/EMRE/MICU1-knockout cells, with MICU1 used to pull down other subunits. C-terminal truncation (ΔC, residues 445–476 deleted) of MICU1 greatly weakens its interaction with MCU without affecting MICU2 binding. Tim23, a membrane-embedded component of the mitochondrial translocase of the inner membrane, was used to rule out nonspecific binding. The bar chart summarizes the effect of MICU1 C-truncation on the gatekeeping function. Wild-type or ΔC MICU1 was expressed in MICU1-knockout cells, and mitochondrial Ca2+ uptake in low-Ca2+ conditions (300 nM) was quantified using 45Ca2+ flux. Results are shown as means ± s.e.m. Numbers of independent repeats are provided in parentheses. ΔC MICU1 has a much weaker ability to gate MCU than wild-type MICU1, as determined by two-tailed t-test (P = 0.0098). Con, untransfected MICU1-knockout cells. e, Roles of the MICU1–EMRE interaction in uniplex stability. The experiment assessed the complex stability of wild-type MCU and the indicated MICU1 constructs in the presence of wild-type or C-truncated (residues 96–107 deleted) EMRE in low-Ca2+ conditions. These three subunits were coexpressed in MCU/MICU1/EMRE-knockout cells. C-truncation of EMRE or charge-reversal of MICU1’s KKKKR sequence to QEQEQ (EQ) greatly weakens MICU1’s association with MCU. f, Contribution of residue R352 to formation of the MICU1–MICU2 heterodimer. Complex formation between C463S MICU1, which cannot form a disulfide MICU dimer, and wild-type or R352E MICU2 was examined in MICU1/MICU2-knockout cells. The R352E mutation in MICU2 strongly perturbs dimerization with MICU1. Letm1, detected using anti-Letm1 antibody, serves as control for nonspecific interactions. MCU and EMRE signals reflect native proteins. All coimmunoprecipitation experiments (c–f) were performed four times with similar results using independent biological samples. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 2 Single-particle cryo-EM analysis of the uniplex in low Ca2+ conditions.
a, Representative cryo-EM image of the purified uniplex in low Ca2+ conditions. b, 2D class averages of the uniplex. c, Workflow for classification and refinement. NU-refinement, non-uniform refinement. d, Angle distributions of the particles for the final reconstruction. e, Fourier shell correlation (FSC) of the final reconstruction as a function of resolution. Red, gold-standard FSC curve, FSC = 0.143; blue, FSC = 0.5; orange, FSC curve between the final model and half-map 1; green, FSC curve between the final model and half-map 2. f, Local resolution of the map calculated by BlocRes.
Extended Data Fig. 3 Representative cryo-EM density maps of the uniplex in low-Ca2+ and high-Ca2+ conditions.
a, b, Cryo-EM density maps of MCU (a) and its selectivity filter (b) for the low-Ca2+ uniplex. The putative cation is shown as a red sphere in b. c, Cryo-EM density map of EMRE of the low-Ca2+ uniplex. d, e, Cryo-EM density of the α-helices in MICU1 (d) and MICU2 (e) from the low-Ca2+ uniplex. f, Cryo-EM density maps of lipids bound to the MCU TM region of the low-Ca2+ uniplex. g–j, Cryo-EM density maps of MCU (g), EMRE (h), MICU1 (i), and MICU2 (j) of the high-Ca2+ uniplex.
Extended Data Fig. 4 Single-particle cryo-EM analysis of the uniplex in high-Ca2+ conditions.
a, Representative cryo-EM image of the uniplex in high-Ca2+ conditions. b, 2D class averages of the uniplex. c, Workflow of classification and refinement. d, Angle distributions of the particles for the final reconstruction. e, The final reconstruction FSC as a function of resolution. Red, gold-standard FSC curve, FSC = 0.143; blue, FSC = 0.5; orange, FSC curve between the final model and half-map 1; green, FSC curve between the final model and half-map 2. f, Local resolution of the map calculated using BlocRes.
Extended Data Fig. 5 Structural comparison of low-Ca2+ uniplex and the MCU–EMRE subcomplex.
a, Structural superposition of the MCU–EMRE part of low-Ca2+ uniplex (blue) and the MCU–EMRE subcomplex (grey). b, Interactions between MCU and EMRE in the uniplex. Two MCU subunits are coloured green and cyan, and one EMRE is coloured orange. c, The selectivity filter of MCU in the uniplex. The side chains of D261 and E264 are shown as sticks. The putative cation is shown as a red sphere. d, Comparison of the luminal gate of MCU in the uniplex (blue) and the MCU–EMRE subcomplex (grey). The cryo-EM density of the uniplex luminal gate is shown on the right. e, Surface representation of the MCU–MICU1 interface, coloured according to electrostatic potential (red, negative; blue, positive).
Extended Data Fig. 6 Structural comparison of the high-Ca2+ uniplex and MCU–EMRE subcomplex.
a, Cryo-EM map of the high-Ca2+ uniplex. b, Superposition of one copy of the high-Ca2+ uniplex (blue) and the MCU–EMRE subcomplex (grey). The MICU1 and MICU2 parts of the uniplex are omitted for clarity. c, Superposition of the dimeric high-Ca2+ uniplex and MCU–EMRE subcomplex. d, Comparison of the luminal gate of MCU in the high-Ca2+ uniplex (blue) and the MCU–EMRE subcomplex (grey). The cryo-EM density of the uniplex luminal gate is shown on the right.
Extended Data Fig. 7 Validation of the interface of the high-Ca2+ uniplex and functional roles of uniplex dimer interfaces.
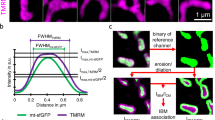
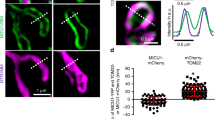
a, The effect of Ca2+ elevation and EMRE C-truncation on MICU1’s association with the uniplex. 1D4-tagged wild-type (WT) MCU was used to precipitate wild-type MICU1 and the indicated EMRE constructs in high- or low-Ca2+ conditions (H and L, respectively). All three subunits were expressed in MCU/MICU1/EMRE-knockout cells. The Letm1 control was performed as in Extended Data Fig. 1c, but with an 1D4-tagged, rather than FLAG-tagged, version of Letm1. Letm1 in WCL or immunoprecipitate (IP) was detected with anti-1D4 or anti-Letm1 antibodies, respectively. Four independent experiments were performed, yielding similar results. b, Roles of MICU1’s polybasic sequence in MICU1 binding to the MCU–EMRE tetramer. The image compares the stability of wild-type MCU complexed with wild-type MICU1 or the KKKKR-to-QEQEQ mutant (EQ) of MICU1 in low or high Ca2+. Wild-type MCU/EMRE and MICU1 constructs were expressed in MCU/MICU1/EMRE-knockout cells. Four independent repeats were performed, leading to similar results. c, Size-exclusion chromatography profiles of the purified human uniplex containing wild-type or D123R MCU. The inset shows SDS–PAGE gel analysis of the uniplex. Data are representative of three independent experiments with similar results. d, Size-exclusion chromatography profiles of the purified human uniplex containing wild-type or mutant MICU2. The uniplex was expressed in MICU2-knockout HEK293 cells to eliminate the effect of endogenous MICU2. The experiment was performed twice independently with similar results. Tetra, R107E–R120E–K121E–D154R mutant MICU2. e–g, Functional roles of the uniporter’s dimer interfaces. We analysed the D123R MCU mutant expressed in MCU-knockout cells, or K121A or R107E–R120E–K121E–D154R (tetra) MICU2 mutants expressed in MICU2-knockout cells, using a standard fluorophore-based mitochondrial Ca2+-uptake assay in 10 μM Ca2+ (e, f) or by 45Ca2+ flux in 300 nM Ca2+ (g). Numbers in parentheses indicate numbers of independent repeats. Arrowheads in e indicate the addition of Ru360. Con, untransfected cells. In 45Ca2+ flux experiments, wild-type MICU1 was coexpressed with wild-type or D123R MCU in MCU-knockout cells to ensure sufficient copies of MICU1 to gate MCU (1 μg MCU and 2 μg MICU1 DNA per well in six-well plates). The tetra-MICU2 construct has lower expression levels despite using threefold more DNA in transient expression. h–j, Localization of the uniporter in the mitochondrial inner membrane of wild-type (h) or MICU2-knockout (KO; j) cells. Mitochondrial membrane fractions enriched in outer membrane, inner/outer-membrane contact points (CP) or inner membrane (IMM) were separated in a sucrose gradient as described54. COX2, mitofilin and VDAC were used as the markers for inner membrane, inner/outer-membrane contact points and outer membrane, respectively. We found MCU to be more enriched in contact points (h, i). This feature was not affected by knocking out MICU2 or expressing tetra-MICU2 in MICU2-knockout cells (i, j). The sucrose gradient goes from 60% to 30% from left to right. The bar chart in i presents the ratio of total western signals in IMM (yellow box in h) over the signals in contact points (cyan box in h). Three biologically independent experiments were carried out, with similar results (h, j), as summarized in the bar chart (i). Two-tailed t-tests were performed, with P-values labelled on the bar chart. k–l, Effect of the D123R mutation on uniporter distribution. D123R or wild-type MCU was expressed in MCU-knockout cells, and MCU localization was analysed. D123R reduces the biased distribution of MCU in contact points. Four biologically independent experiments were performed, with similar results (k), as summarized in the bar chart (l). Statistical analyses were carried out through two-tailed t-tests. All bar charts (f, g, i, l) present data as means ± s.e.m. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 8 MICU1–MICU2 heterodimer in low-Ca2+ and high-Ca2+ conditions and sequence alignment of MICU2.
a, Structural comparison of canonical EF hands 1 and 4 of MICU1 in the low-Ca2+ uniplex (green) with those known in the Ca2+-free (grey) and Ca2+-bound (orange) states. b, Structural comparison of canonical EF hands 1 and 4 of MICU2 in the low-Ca2+ uniplex (cyan) versus those known in the Ca2+-free (wheat) and Ca2+-bound (yellow) states. c, Structural comparison of canonical EF hands 1 and 4 of MICU1 in the high-Ca2+ uniplex (lemon) with those known in the Ca2+-bound (orange) and Ca2+-free (grey) states. d, Structural comparison of canonical EF hands 1 and 4 of MICU2 in the high-Ca2+ uniplex (blue) versus those known in the Ca2+-bound (yellow) and Ca2+-free (wheat) states. e, Sequence alignment of MICU2 homologues from Homo sapiens (Hs), Mus musculus (Mm), Gallus gallus (Gg), Xenopus tropicalis (Xt) and Danio rerio (Dr). The residues participating in MICU1–MICU2 interactions are indicated with magenta circles.
Extended Data Fig. 9 Sequence alignment of MICU1 from different species.
Sequence alignment of MICU1 homologues from Homo sapiens (Hs), Gallus gallus (Gg), Danio rerio (Dr), Ciona intestinalis (Ci), Drosophila melanogaster (Dm), Caenorhabditis elegans (Ce), Arabidopsis thaliana (At) and Dictyostelium discoideum (Dd). The residues participating in MICU1–MICU2 and MICU1–MCU interactions are indicated with magenta and blue circles, respectively.
Supplementary information
Supplementary Figure 1
Source images for gel electrophoresis and western blot in Figure 2c, Extended Data Figure 1 and Extended Data Figure 7.
Rights and permissions
About this article
Cite this article
Fan, M., Zhang, J., Tsai, CW. et al. Structure and mechanism of the mitochondrial Ca2+ uniporter holocomplex. Nature 582, 129–133 (2020). https://doi.org/10.1038/s41586-020-2309-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2309-6
This article is cited by
-
Targeting paraptosis in cancer: opportunities and challenges
Cancer Gene Therapy (2024)
-
Construction and in vitro evaluation of pH-sensitive nanoparticles to reverse drug resistance of breast cancer stem cells
Discover Oncology (2024)
-
Mitochondrial dysfunction: roles in skeletal muscle atrophy
Journal of Translational Medicine (2023)
-
Reduced mitochondrial calcium uptake in macrophages is a major driver of inflammaging
Nature Aging (2023)
-
A Treg-specific long noncoding RNA maintains immune-metabolic homeostasis in aging liver
Nature Aging (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.