Abstract
Acetaldehyde is a highly reactive, DNA-damaging metabolite that is produced upon alcohol consumption1. Impaired detoxification of acetaldehyde is common in the Asian population, and is associated with alcohol-related cancers1,2. Cells are protected against acetaldehyde-induced damage by DNA crosslink repair, which when impaired causes Fanconi anaemia (FA), a disease resulting in failure to produce blood cells and a predisposition to cancer3,4. The combined inactivation of acetaldehyde detoxification and the FA pathway induces mutation, accelerates malignancies and causes the rapid attrition of blood stem cells5,6,7. However, the nature of the DNA damage induced by acetaldehyde and how this is repaired remains a key question. Here we generate acetaldehyde-induced DNA interstrand crosslinks and determine their repair mechanism in Xenopus egg extracts. We find that two replication-coupled pathways repair these lesions. The first is the FA pathway, which operates using excision—analogous to the mechanism used to repair the interstrand crosslinks caused by the chemotherapeutic agent cisplatin. However, the repair of acetaldehyde-induced crosslinks results in increased mutation frequency and an altered mutational spectrum compared with the repair of cisplatin-induced crosslinks. The second repair mechanism requires replication fork convergence, but does not involve DNA incisions—instead the acetaldehyde crosslink itself is broken. The Y-family DNA polymerase REV1 completes repair of the crosslink, culminating in a distinct mutational spectrum. These results define the repair pathways of DNA interstrand crosslinks caused by an endogenous and alcohol-derived metabolite, and identify an excision-independent mechanism.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Datasets generated during the current study are available from the corresponding authors upon reasonable request.
Code availability
Code used in the current study is available from the corresponding authors upon reasonable request.
References
Brooks, P. J., Enoch, M. A., Goldman, D., Li, T. K. & Yokoyama, A. The alcohol flushing response: an unrecognized risk factor for esophageal cancer from alcohol consumption. PLoS Med. 6, e1000050 (2009).
Lai, C. L. et al. Dominance of the inactive Asian variant over activity and protein contents of mitochondrial aldehyde dehydrogenase 2 in human liver. Alcohol. Clin. Exp. Res. 38, 44–50 (2014).
Langevin, F., Crossan, G. P., Rosado, I. V., Arends, M. J. & Patel, K. J. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature 475, 53–58 (2011).
Garaycoechea, J. I. & Patel, K. J. Why does the bone marrow fail in Fanconi anemia? Blood 123, 26–34 (2014).
Hira, A. et al. Variant ALDH2 is associated with accelerated progression of bone marrow failure in Japanese Fanconi anemia patients. Blood 122, 3206–3209 (2013).
Garaycoechea, J. I. et al. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature 489, 571–575 (2012).
Garaycoechea, J. I. et al. Alcohol and endogenous aldehydes damage chromosomes and mutate stem cells. Nature 553, 171–177 (2018).
Wang, M. et al. Identification of DNA adducts of acetaldehyde. Chem. Res. Toxicol. 13, 1149–1157 (2000).
Cho, Y. J. et al. Stereospecific formation of interstrand carbinolamine DNA cross-links by crotonaldehyde- and acetaldehyde-derived α-CH3-γ-OH-1,N 2-propano-2′-deoxyguanosine adducts in the 5′-CpG-3′ sequence. Chem. Res. Toxicol. 19, 195–208 (2006).
Loudon, G. M. Organic Chemistry 874–878 (Oxford Univ. Press, 2002).
Duxin, J. P. & Walter, J. C. What is the DNA repair defect underlying Fanconi anemia? Curr. Opin. Cell Biol. 37, 49–60 (2015).
Knipscheer, P. et al. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science 326, 1698–1701 (2009).
Long, D. T., Räschle, M., Joukov, V. & Walter, J. C. Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science 333, 84–87 (2011).
Räschle, M. et al. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell 134, 969–980 (2008).
Semlow, D. R., Zhang, J., Budzowska, M., Drohat, A. C. & Walter, J. C. Replication-dependent unhooking of DNA interstrand cross-links by the NEIL3 glycosylase. Cell 167, 498–511 (2016).
Duxin, J. P., Dewar, J. M., Yardimci, H. & Walter, J. C. Repair of a DNA-protein crosslink by replication-coupled proteolysis. Cell 159, 346–357 (2014).
Zhang, J. et al. DNA interstrand cross-link repair requires replication-fork convergence. Nat. Struct. Mol. Biol. 22, 242–247 (2015).
Klein Douwel, D. et al. XPF-ERCC1 acts in unhooking DNA interstrand crosslinks in cooperation with FANCD2 and FANCP/SLX4. Mol. Cell 54, 460–471 (2014).
Budzowska, M., Graham, T. G. W., Sobeck, A., Waga, S. & Walter, J. C. Regulation of the Rev1–Pol ζ complex during bypass of a DNA interstrand cross-link. EMBO J. 34, 1971–1985 (2015).
Johnson, R. E., Washington, M. T., Haracska, L., Prakash, S. & Prakash, L. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature 406, 1015–1019 (2000).
Matsuda, T., Kawanishi, M., Yagi, T., Matsui, S. & Takebe, H. Specific tandem GG to TT base substitutions induced by acetaldehyde are due to intra-strand crosslinks between adjacent guanine bases. Nucleic Acids Res. 26, 1769–1774 (1998).
Huang, H., Zhu, L., Reid, B. R., Drobny, G. P. & Hopkins, P. B. Solution structure of a cisplatin-induced DNA interstrand cross-link. Science 270, 1842–1845 (1995).
Cho, Y. J., Kozekov, I. D., Harris, T. M., Rizzo, C. J. & Stone, M. P. Stereochemistry modulates the stability of reduced interstrand cross-links arising from R- and S-α-CH3-γ-OH-1,N 2-propano-2′-deoxyguanosine in the 5′-CpG-3′ DNA sequence. Biochemistry 46, 2608–2621 (2007).
Amunugama, R. et al. Replication fork reversal during DNA interstrand crosslink repair requires CMG unloading. Cell Rep. 23, 3419–3428 (2018).
Huang, J. et al. The DNA translocase FANCM/MHF promotes replication traverse of DNA interstrand crosslinks. Mol. Cell 52, 434–446 (2013).
Acknowledgements
We thank G. P. Crossan, J. T. P. Yeeles and members of the Knipscheer and Patel laboratories for critical reading of the manuscript; Hubrecht animal caretakers for animal support; J. C. Walter and D. R. Semlow for providing us with recombinant X. laevis NEIL3 proteins, the AP-ICL and lacO plasmids; and S. Y. Peak-Chew, S. Maslen and M. Skehel for mass spectrometry analysis. This work was supported by a project grant from the Dutch Cancer Society (KWF HUBR 2015-7736 to P.K.), the Gravitation program CancerGenomiCs.nl from the Netherlands Organisation for Scientific Research (NWO), part of the Oncode Institute, which is partly financed by the Dutch Cancer Society. K.S. was supported by the Uehara Memorial Foundation, the Mochida Memorial Foundation for Medical and Pharmaceutical Research, and the JSPS Postdoctoral Fellowship for Research Abroad. A.N.K.-L. was supported by the Wellcome Trust. J.S. was supported by Cancer Research UK. This work was supported by a European Research Council Starting Grant (ERC-STG 678423-EpiID) to J.K. and K.R. and an NWO Veni grant (016.Veni.181.013) to K.R.
Author information
Authors and Affiliations
Contributions
K.J.P. and P.K. initiated and supervised the study. M.R.H. co-ordinated key aspects of the project. K.S., A.N.K.-L. and K.R. contributed equally. M.R.H., M.M., J.S., M.P., D.M.W. and J.W.C. designed the strategy and synthesized the AA-ICL and PdG adducts. A.N.K.-L. performed cell toxicity experiments. A.B., M.W. and P.K. designed biochemical assays in Xenopus egg extracts. A.B., K.S. and M.W. conducted Xenopus extract assays. K.R. and J.K. performed bioinformatic analysis. K.J.P. and P.K. wrote the manuscript and M.R.H. designed and created the figures.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
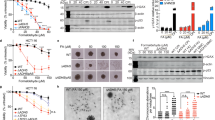
Extended Data Fig. 1 Acetaldehyde ICLs are stable and are repaired in Xenopus egg extracts.
a, Scheme for the synthesis of the precursor, 4-(R)-aminopentane-1,2-diol. b, Site-specific synthesis of a PdG adduct in a DNA oligonucleotide. c, Denaturing PAGE showing crosslink formation between dG and PdG, but not between dG and inosine (ino), which lacks an N2 amine. Two independent experiments were performed. d, Confirmation of AA-ICL formation by MALDI MS. The peak at m/z 12,370.2 represents the imine or pyrimidopurinone form. Two further peaks at m/z 5,979.41 and 6,409.74 equate to masses for the two parent oligonucleotides, consistent with the mass of the carbinolamine form after dissociation back to PdG and dG under the desorption/ionization conditions. Three independent experiments were performed. e, Stability of AANAT-ICL as a function of temperature and time, as determined by radiolabelling ([α-32P]dCTP) and resolution by denaturing PAGE. Error bars represent s.e.m. from three independent experiments. f, AANAT-ICL is susceptible to hydrolysis in aqueous acid, whereas AARED-ICL is stable. Pre-purification crosslink reactions were incubated with or without formic acid and products were resolved by denaturing PAGE. Three independent experiments were performed. g, Scheme depicting the type and position of the DNA lesions used in this study. Duplex DNA with or without the indicated lesions was annealed into a backbone vector to generate circular plasmids with or without damage. h, To determine the percentage of crosslinks, the ICL-containing plasmids were digested with NotI, labelled at the 3′-end by end filling with [α-32P]dCTP, and separated by denaturing PAGE. Crosslinked DNA (88 nt) shows slower mobility compared with non-crosslinked DNA (44 nt). The percentage of crosslinks was calculated by comparing the 88-nt product with the 44-nt products. Two independent experiments were performed. i, AANAT-ICL and Pt-ICL are stable in Xenopus egg extracts. Plasmids were incubated in a high-speed supernatant (HSS) extract. DNA was extracted and analysed as described in h. Three independent experiments were performed. j, Solution structures of a cisplatin ICL and a reduced form of an acetaldehyde ICL (PDB: 1DDP22 and 2HMD23, cartoon representation generated in PyMOL). k, The indicated plasmids were replicated in Xenopus egg extracts and repair intermediates were digested with NotI, labelled at the 3′-end, and resolved by denaturing PAGE. The increase in intensity of the 44-nt band over time indicates ongoing replication and repair. A higher mobility band, probably generated from end-joining activity in some extracts, is indicated by an asterisk. This gel is the independent experimental duplicate of that in Fig. 1e. l, Quantification of repair based on the intensity of the 44-nt product on the gel in k, as described in the Supplementary Methods. This graph is the independent experimental duplicate of that in Fig. 1f. Additional replicates of these experiments are presented in Fig. 2a, Extended Data Figs. 2k–m, 4b.
Extended Data Fig. 2 Acetaldehyde ICLs are repaired by Fanconi-dependent and Fanconi-independent mechanisms.
a, Model for ICL repair by the FA pathway. Upon convergence of two replication forks at the crosslink, the CMG helicase is unloaded from the DNA to enable the approach of one replication fork to the −1 position. Ubiquitylation of FANCD2 promotes the recruitment of the XPF-ERCC1-SLX4 (XES) complex to the ICL, which enables nucleolytic incisions that unhook the crosslink. This step could be preceded by fork reversal of one of the stalled replication forks24. Incisions generate a broken strand and a strand with an adduct; the latter is bypassed by TLS whereas the broken strand is repaired by homologous recombination. In mammalian cells, it has been shown that a single fork can pass over the ICL without unhooking25; this ‘traverse’ gives rise to a structure that resembles the one generated after fork convergence and CMG unloading and could follow the same steps subsequently. b, The indicated plasmids were replicated in Xenopus egg extracts and reaction samples were analysed by western blot with FANCD2 antibody. Two independent experiments were performed. c, Western blot of FANCD2, showing a titration of Xenopus egg extracts compared to mock and FANCD2-depleted extracts. Two independent experiments were performed. d, The indicated plasmids were replicated in mock or in FANCD2-depleted extracts in the presence of [α-32P]dCTP. Repair products were digested by AflIII, separated on a sequencing gel alongside a ladder derived from extension primer S, and visualized by autoradiography. The white arrow denotes the −1 product, which is 2 nt larger in pICL-Pt owing to the position of the ICL. Three independent experiments were performed. e, The indicated plasmids were replicated in mock or FANCD2-depleted extracts and repair intermediates were digested with NotI, labelled at the 3′-end, and resolved by denaturing PAGE. Quantification of repair based on the intensity of the 44-nt product is shown in Fig. 1g. f, The independent experimental duplicate of Fig. 1g. g, The independent experimental triplicate of Fig. 1g, but using only pICL-AANAT. h, Plasmid pICL-AANAT was replicated in FANCD2-depleted extract, or FANCD2-depleted extract supplemented with a recombinant FANCI–FANCD2 complex (ID). Reaction samples were resolved by native agarose gel and visualized by autoradiography. RRIs, open circle (OC) and supercoiled (SC) products are indicated. The stalled repair product (grey arrow) is indicated. Two independent experiments were performed. i, The indicated plasmids were replicated in Xenopus egg extracts in the presence or in the absence of p97i and the intermediates were resolved by native agarose gel electrophoresis. The stalled repair products (grey arrow) are indicated. Seven independent experiments were performed. j, The indicated plasmids were replicated in Xenopus egg extracts in the presence or in the absence of p97i, and repair intermediates were digested with NotI, labelled at the 3′-end, and resolved by denaturing PAGE. The increase in intensity of the 44-nt band (white arrow) over time indicates ongoing replication and repair. A higher mobility band, probably generated from end-joining activity in some extracts, is indicated with an asterisk. k, Quantification of repair based on the intensity of the 44-nt product on the gel shown in j, as indicated in the Supplementary Methods. l, Quantified independent experimental duplicate of j. m, Quantified independent experimental triplicate of j.
Extended Data Fig. 3 Reduced acetaldehyde ICLs are repaired by the Fanconi pathway.
a, MALDI MS data confirms the identity and the stability of the reduced ICL. Three independent experiments were performed. b, Plasmids were replicated in extract and products were resolved on a native agarose gel. Three independent experiments were performed. c, The indicated plasmids were replicated in Xenopus egg extracts in the presence or in the absence of p97i and the repair intermediates were analysed using the NotI digestion assay. The increase in intensity of the 44-nt band indicates ongoing replication and repair. A band with higher mobility, probably generated from end-joining activity in some extracts, is indicated with an asterisk. Quantification of repair based on this gel is shown in Fig. 1h. Three independent experiments were performed. d, The independent experimental duplicate of Fig. 1h. e, The independent experimental triplicate of Fig. 1h.
Extended Data Fig. 4 Acetaldehyde ICL repair requires DNA replication and replication fork convergence.
a, The indicated plasmids were replicated in Xenopus egg extracts in the presence or in the absence of geminin. Repair intermediates were digested with NotI, labelled at the 3′-end, and resolved by denaturing PAGE. Quantification of repair based on the intensity of the 44-nt product is shown in Fig. 2a. b, The independent experimental duplicate of Fig. 2a. c, The independent experimental triplicate of Fig. 2a but using only pICL-AANAT. d, pICL-Pt-lacO was replicated in Xenopus egg extracts containing [α-32P]dCTP in the presence or in the absence of LacR. The repair intermediates were digested by AflIII and EcoRI, separated on a sequencing gel and visualized by autoradiography. Two independent experiments were performed. e, The indicated plasmids were replicated in extracts in the presence or in the absence of LacR. Repair products were digested with NotI, labelled at the 3′-end, and resolved by denaturing PAGE. Three independent experiments were performed. f, Quantification of repair based on the intensity of the 44-nt product on the gel shown in e, as described in the Supplementary Methods. Three independent experiments were performed. g, The independent experimental duplicate of f. h, The independent experimental triplicate of f.
Extended Data Fig. 5 The alternative route of AA-ICL repair does not involve DNA excision.
a, Western blot showing a titration of Xenopus egg extracts compared to the NEIL3-depleted (∆NEIL3) extract and NEIL3-depleted extract supplemented with recombinant wild-type NEIL3 (WT) or catalytically inactive NEIL3 (MUT). Three independent experiments were performed. b, The indicated plasmids were replicated in NEIL3-depleted extract containing [α-32P]dCTP, supplemented with wild-type (WT) or catalytically inactive (MUT) NEIL3. Replication intermediates were resolved by native agarose gel electrophoresis and visualized by autoradiography. Three independent experiments were performed. c, Clonogenic survival of wild-type, NEIL3-, FANCL- or NEIL3/FANCL-deficient human HAP1 cells after a 2-h exposure to acetaldehyde. Three independent experiments were performed. Data are mean ± s.e.m. d, The median lethal dose (LD50) of acetaldehyde for the survival of wild-type and deficient HAP1 cells was calculated by regression analysis of the curves presented in c. Data are mean ± s.e.m. Three independent experiments were performed. e, Quantification of the arm fragments resulting from APE1 treatment (AP sites) from the gel in Fig. 2e. f, Quantification of the APE1 arms as in e, from an independent duplicate experiment without the addition of p97i. g, Quantification of the APE1 arms as in e, from an independent triplicate experiment without the addition of p97i. As a positive control we used a plasmid containing an abasic-site-induced interstrand crosslink (pICL-AP) that is also repaired via the glycosylase NEIL3. h, Quantification of the HincII arm fragments from the gel in Fig. 2f. i, Quantification of the HincII arms as in h, from an independent duplicate experiment. j, Quantification of the HincII arms as in h, from an independent triplicate experiment. k, Schematic representation of the formation of DNA adducts by unhooking incisions during ICL repair (left). These adducts are not removed during ICL repair in Xenopus egg extracts14 and can therefore be visualized. Plasmids were replicated in Xenopus egg extracts in the presence or in the absence of p97i. Late reaction samples were digested with AflIII and AseI and separated on a sequencing gel. Adducts on either the top or the bottom strand (white arrowheads) were detected by strand-specific Southern blotting (right). Three independent experiments were performed.
Extended Data Fig. 6 Both routes of AA-ICL repair are mediated by REV1 and REV7.
a, The indicated plasmids were replicated in mock or REV1-depleted extracts, in the presence or in the absence of p97i. Reaction intermediates were digested by either AflIII or AflIII and BamHI, separated on a sequencing gel alongside a ladder derived from extension of primer S, and visualized by autoradiography. White arrows denote 0 products, dark grey arrows indicate −1 products, and light grey arrows indicate 0/−1 products (not separated). Two independent experiments were performed. b, Western blot detection of REV7 in REV1- or mock-depleted Xenopus egg extracts compared to a titration of undepleted extract. Two independent experiments were performed. c, Western blot detection of REV7 and REV1 in REV7-depleted (∆REV7) or mock-depleted Xenopus egg extracts compared to a titration of undepleted extract. Two independent experiments were performed. d, The indicated plasmids were replicated in mock- or REV7-depleted extracts and reaction intermediates were digested by either AflIII or AflIII and BamHI, separated on a sequencing gel and visualized by autoradiography. Grey arrows indicate −1 products. Two independent experiments were performed. e, Indicated plasmids were replicated in mock- or REV1-depleted extracts and reaction intermediates were digested by AflIII and BamHI, separated on a sequencing gel alongside a ladder derived from extension of primer S, and visualized by autoradiography. The asterisk indicates a 121-nt background fragment caused by a second BamHI restriction site in the leftward fork. Two independent experiments were performed.
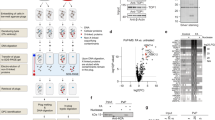
Extended Data Fig. 7 Mutagenic outcome of acetaldehyde crosslink repair.
a, Frequency of nucleotide misincorporation in a 15-bp region flanking the lesions present in the indicated plasmids. The mutation frequencies for the same plasmid that has not been replicated in Xenopus egg extracts (NR) and for the control vector (pCon) are plotted together. Strand specificity is lost because the sample preparation involves PCR amplification; as such, only the top sequence is indicated below the graphs. See Fig. 4. b, Distribution and frequency of nucleotide misincorporations in a 15-bp region flanking the lesions present in the indicated plasmids. Independent duplicate sequencing experiment of Fig. 4b. The heights of the bars represent the mutation frequency minus the baseline mutations found in pCon. c, Frequency of nucleotide misincorporations in a 15-bp region flanking the lesions present in the indicated plasmids (data from the same sequencing experiment as in b). The mutation frequency for pCon is also plotted.
Supplementary information
Supplementary Information
This file contains Supplementary Notes 1-3 with a short guide.
Rights and permissions
About this article
Cite this article
Hodskinson, M.R., Bolner, A., Sato, K. et al. Alcohol-derived DNA crosslinks are repaired by two distinct mechanisms. Nature 579, 603–608 (2020). https://doi.org/10.1038/s41586-020-2059-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2059-5
This article is cited by
-
Ethanol Causes Cell Death and Neuronal Differentiation Defect During Initial Neurogenesis of the Neural Retina by Disrupting Calcium Signaling in Human Retinal Organoids
Stem Cell Reviews and Reports (2023)
-
Acetaldehyde induces similar cytotoxic and genotoxic risks in BEAS-2B cells and HHSteCs: involvement of differential regulation of MAPK/ERK and PI3K/AKT pathways
Environmental Science and Pollution Research (2023)
-
Polyphenols from persimmon fruit attenuate acetaldehyde-induced DNA double-strand breaks by scavenging acetaldehyde
Scientific Reports (2022)
-
The structure-specific endonuclease complex SLX4–XPF regulates Tus–Ter-induced homologous recombination
Nature Structural & Molecular Biology (2022)
-
Uncovering cancer vulnerabilities by machine learning prediction of synthetic lethality
Molecular Cancer (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.