Abstract
Mitochondrial fidelity is tightly linked to overall cellular homeostasis and is compromised in ageing and various pathologies1,2,3. Mitochondrial malfunction needs to be relayed to the cytosol, where an integrated stress response is triggered by the phosphorylation of eukaryotic translation initiation factor 2α (eIF2α) in mammalian cells4,5. eIF2α phosphorylation is mediated by the four eIF2α kinases GCN2, HRI, PERK and PKR, which are activated by diverse types of cellular stress6. However, the machinery that communicates mitochondrial perturbation to the cytosol to trigger the integrated stress response remains unknown1,2,7. Here we combine genome engineering and haploid genetics to unbiasedly identify genes that affect the induction of C/EBP homologous protein (CHOP), a key factor in the integrated stress response. We show that the mitochondrial protease OMA1 and the poorly characterized protein DELE1, together with HRI, constitute the missing pathway that is triggered by mitochondrial stress. Mechanistically, stress-induced activation of OMA1 causes DELE1 to be cleaved into a short form that accumulates in the cytosol, where it binds to and activates HRI via its C-terminal portion. Obstruction of this pathway can be beneficial or adverse depending on the type of mitochondrial perturbation. In addition to the core pathway components, our comparative genetic screening strategy identifies a suite of additional regulators. Together, these findings could be used to inform future strategies to modulate the cellular response to mitochondrial dysfunction in the context of human disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
All data including Source Data for Figs. 1–3 and Extended Data Fig. 1, 5–11 are provided with the paper and its Supplementary Information files. Deep-sequencing raw data (genome-wide genetic screens and RNA-seq) have been deposited in the NCBI Sequence Read Archive under accession number PRJNA559719. The corresponding processed data are provided in Supplementary Tables 1–3, 6–10. Materials and reagents are available from the corresponding author on request.
References
Higuchi-Sanabria, R., Frankino, P. A., Paul, J. W. III, Tronnes, S. U. & Dillin, A. A futile battle? Protein quality control and the stress of aging. Dev. Cell 44, 139–163 (2018).
Shpilka, T. & Haynes, C. M. M. The mitochondrial UPR: mechanisms, physiological functions and implications in ageing. Nat. Rev. Mol. Cell Biol. 19, 109–120 (2018).
Sun, N., Youle, R. J. & Finkel, T. The mitochondrial basis of aging. Mol. Cell 61, 654–666 (2016).
Quirós, P. M. et al. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J. Cell Biol. 216, 2027–2045 (2017).
Harding, H. P. et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11, 619–633 (2003).
Taniuchi, S., Miyake, M., Tsugawa, K., Oyadomari, M. & Oyadomari, S. Integrated stress response of vertebrates is regulated by four eIF2α kinases. Sci. Rep. 6, 32886 (2016).
Münch, C. & Harper, J. W. Mitochondrial unfolded protein response controls matrix pre-RNA processing and translation. Nature 534, 710–713 (2016).
Palam, L. R., Baird, T. D. & Wek, R. C. Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J. Biol. Chem. 286, 10939–10949 (2011).
Brockmann, M. et al. Genetic wiring maps of single-cell protein states reveal an off-switch for GPCR signalling. Nature 546, 307–311 (2017).
Azim, M. & Surani, H. Glycoprotein synthesis and inhibition of glycosylation by tunicamycin in preimplantation mouse embryos: compaction and trophoblast adhesion. Cell 18, 217–227 (1979).
Yore, M. M., Kettenbach, A. N., Sporn, M. B., Gerber, S. A. & Liby, K. T. Proteomic analysis shows synthetic oleanane triterpenoid binds to mTOR. PLoS One 6, e22862 (2011).
Harada, T., Iwai, A. & Miyazaki, T. Identification of DELE, a novel DAP3-binding protein which is crucial for death receptor-mediated apoptosis induction. Apoptosis 15, 1247–1255 (2010).
Tanaka, A. et al. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 191, 1367–1380 (2010).
Baker, M. J. et al. Stress-induced OMA1 activation and autocatalytic turnover regulate OPA1-dependent mitochondrial dynamics. EMBO J. 33, 578–593 (2014).
Sekine, S. et al. Reciprocal roles of Tom7 and OMA1 during mitochondrial import and activation of PINK1. Mol. Cell 73, 1028–1043 (2019).
Scheufler, C. et al. Structure of TPR domain–peptide complexes: critical elements in the assembly of the Hsp70–Hsp90 multichaperone machine. Cell 101, 199–210 (2000).
Lu, L., Han, A. P. & Chen, J.-J. J. Translation initiation control by heme-regulated eukaryotic initiation factor 2α kinase in erythroid cells under cytoplasmic stresses. Mol. Cell. Biol. 21, 7971–7980 (2001).
Shah, D. I. et al. Mitochondrial Atpif1 regulates haem synthesis in developing erythroblasts. Nature 491, 608–612 (2012).
Sukumar, M. et al. Mitochondrial membrane potential identifies cells with enhanced stemness for cellular therapy. Cell Metab. 23, 63–76 (2016).
Baricault, L. et al. OPA1 cleavage depends on decreased mitochondrial ATP level and bivalent metals. Exp. Cell Res. 313, 3800–3808 (2007).
Sorrentino, V. et al. Enhancing mitochondrial proteostasis reduces amyloid-β proteotoxicity. Nature 552, 187–193 (2017).
Sidrauski, C. et al. Pharmacological brake-release of mRNA translation enhances cognitive memory. eLife 2, e00498 (2013).
Züchner, S. et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot–Marie–Tooth neuropathy type 2A. Nat. Genet. 36, 449–451 (2004).
Dickson, M. A. et al. Human keratinocytes that express hTERT and also bypass a p16INK4a-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol. Cell. Biol. 20, 1436–1447 (2000).
Schmid-Burgk, J. L., Höning, K., Ebert, T. S. & Hornung, V. CRISPaint allows modular base-specific gene tagging using a ligase-4-dependent mechanism. Nat. Commun. 7, 12338 (2016).
Jae, L. T. et al. Lassa virus entry requires a trigger-induced receptor switch. Science 344, 1506–1510 (2014).
Carette, J. E. et al. Haploid genetic screens in human cells identify host factors used by pathogens. Science 326, 1231–1235 (2009).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Schmitt, S. et al. A semi-automated method for isolating functionally intact mitochondria from cultured cells and tissue biopsies. Anal. Biochem. 443, 66–74 (2013).
Bradford, M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976).
Zamzami, N., Métivier, D. & Kroemer, G. Quantitation of mitochondrial transmembrane potential in cells and in isolated mitochondria. Methods Enzymol. 322, 208–213 (2000).
Zischka, H. et al. Electrophoretic analysis of the mitochondrial outer membrane rupture induced by permeability transition. Anal. Chem. 80, 5051–5058 (2008).
Afgan, E. et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537–W544 (2018).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
de Hoon, M. J. L., Imoto, S., Nolan, J. & Miyano, S. Open source clustering software. Bioinformatics 20, 1453–1454 (2004).
Saldanha, A. J. Java Treeview—extensible visualization of microarray data. Bioinformatics 20, 3246–3248 (2004).
Han, H. et al. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 46, D380–D386 (2018).
Bao, X. R. et al. Mitochondrial dysfunction remodels one-carbon metabolism in human cells. eLife 5, e10575 (2016).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000).
The Gene Ontology Consortium. The Gene Ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 47, D330–D338 (2019).
Carbon, S. et al. AmiGO: online access to ontology and annotation data. Bioinformatics 25, 288–289 (2009).
Quirós, P. M., Langer, T. & López-Otín, C. New roles for mitochondrial proteases in health, ageing and disease. Nat. Rev. Mol. Cell Biol. 16, 345–359 (2015).
Calvo, S. E., Clauser, K. R. & Mootha, V. K. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 44, D1251–D1257 (2016).
Acknowledgements
We thank A. Graf and H. Blum for deep-sequencing infrastructure; C. Jung for assistance with confocal microscopy; N. Tafrishi for assistance with cell sorting; S. Theurich for access to Seahorse instrumentation; T. Brummelkamp and V. Hornung for valuable comments on the manuscript; and J. Stingele, R. Beckmann and all members of the Jae lab for helpful discussions. This work was supported by ERC StG 804182 (SOLID), the Center for Integrated Protein Science Munich and the German Research Foundation (Heinz-Maier-Leibnitz Prize) to L.T.J.
Author information
Authors and Affiliations
Contributions
E.F. and L.T.J. conceived the study; E.F., E.-M.E. and L.T.J. performed experiments and analysed data; S.S. performed mitochondrial isolations and subsequent in vitro stimulations; I.A.M. generated knockout cell lines; M.F.M.-B. performed bioinformatics analyses; J.P.-M. generated and analysed RNA-seq libraries; S.K. performed deep sequencing; M.H. constructed plasmids; H.Z. and L.T.J. supervised experiments; E.F. and L.T.J. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Ulrich Elling, Cole Haynes and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 CHOP and CHOPNeon protein levels in the context of different types of pharmacological stimulation.
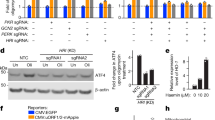
a, Wild-type HAP1 cells were treated as indicated for 9 h and analysed by immunoblotting. TM, tunicamycin. b, Schematic depicting the expected cellular activities of CCCP, tunicamycin and CDDO. ER, endoplasmic reticulum; LONP, LON protease 1; ΔΨ, mitochondrial membrane potential. c, CRISPR engineering of the DDIT3 locus, resulting in an endogenous in-frame fusion of CHOP with a triple Flag tag followed by mNeon as indicated. d, Pharmacological stimulation of wild-type HAP1 CHOPNeon cells for 9 h (tunicamycin and CDDO) or 16 h (CCCP) leads to induction of the CHOPNeon protein, which was measured by flow cytometry (one representative experiment shown of three independent experiments). e, Schematic depicting the generation of CHOPNeon cells and their interrogation in phenotypic genetic screening after exposure to cellular stress. Mutagenized cells were sorted on the basis of mNeon intensity (NeonLo and NeonHi populations) and gene-trap mutations in these populations were analysed by deep sequencing. f, Regulators of CHOPNeon in the genome-wide screen using CDDO (n = 1.90 × 107 interrogated single cells). Genes are coloured as in Fig. 1a, b, with dark grey denoting significant enrichment for mutations (two-sided Fisher’s exact test, FDR-corrected P value (Padj) < 0.05). The two other eIF2α kinases GCN2 (encoded by EIF2AK4) and PKR (encoded by EIF2AK2) are also highlighted in black. g, HAP1 cells were exposed to sgRNAs that target one of the four eIF2α kinases, pharmacologically stimulated for 9 h as indicated and assayed for CHOP induction by immunoblotting (one representative experiment shown of two independent experiments). h–m, Common regulators of CHOPNeon in the genome-wide screens using CCCP, tunicamycin or CDDO. Data are from Fig. 1a, b and panel f, with the statistical significance of mutation enrichment assessed and visualized in the same way. Genes that were identified as significant positive regulators (Padj < 0.05) with a mutation ratio lower than 0.25 (h–j) or negative regulators with a mutation ratio higher than 4 (k–m) and which are shared across all three datasets are highlighted in red. WHSC2 is also known as NELFA.
Extended Data Fig. 2 Unique negative regulators of CHOPNeon across genetic screens.
a–c, Data from Fig. 1a, b and Extended Data Fig. 1f, with statistical significance of mutation enrichment assessed and visualized in the same way. Genes that were identified as significant negative regulators (two-sided Fisher’s exact test, Padj < 0.05) and which had a mutation ratio higher than 4 in the query dataset but not in either of the other two datasets are highlighted in red: CCCP versus tunicamycin and CDDO (a); tunicamycin versus CDDO and CCCP (b); CDDO versus CCCP and tunicamycin (c). ATP5B is also known as ATP5F1B; ATP5F1 as ATP5PB; ATP5O as ATP5PO; GNB2L1 as RACK1; C2orf29 as CNOT11; HIST1H4D as H4C4; and ICT1 as MRPL58.
Extended Data Fig. 3 Unique positive regulators of CHOPNeon across genetic screens.
a–c, Data from Fig. 1a, b and Extended Data Fig. 1f, with statistical significance of mutation enrichment assessed and visualized in the same way. Genes that were identified as significant positive regulators (two-sided Fisher’s exact test, Padj < 0.05) and which had a mutation ratio lower than 0.25 in the query dataset but not in either of the other two datasets are highlighted in red: CCCP versus tunicamycin and CDDO (a); tunicamycin versus CDDO and CCCP (b); CDDO versus CCCP and tunicamycin (c). ATPIF1 is also known as ATP5IF1; KIAA0141 is also known as DELE1; KIAA0664 is also known as CLUH; TH1L is also known as NELFCD; C11orf73 is also known as HIKESHI; C22orf29 is also known as RTL10; H2AFZ is also known as H2AZ1.
Extended Data Fig. 4 Overview of positive and negative regulators of CHOPNeon after treatment with different drugs.
a–c, Data from Fig. 1a, b and Extended Data Fig. 1f, with statistical significance of mutation enrichment assessed and visualized in the same way. For all datasets, significant positive regulators (two-sided Fisher’s exact test, Padj < 0.05) with a mutation ratio lower than 0.25 (corresponding to an effect size of 4× or greater) are shown in blue, whereas significant negative regulators (two-sided Fisher’s exact test, Padj < 0.05) with a mutation ratio higher than 4 are shown in yellow.
Extended Data Fig. 5 Intersection of the most pronounced positive regulators of CHOPNeon across genetic screens and validation of DELE1 and HRI as mediators of mitochondrial stress.
a, Data from Fig. 1a, b and Extended Data Fig. 1f, processed in the same way, except that genes that were identified as highly significant positive regulators (two-sided Fisher’s exact test, Padj < 10−5) with a mutation ratio lower than 0.25 in the CCCP dataset but not in either of the other two datasets are highlighted in red. b, Venn diagram showing the results of stringent filtering of positive CHOPNeon regulators from a (two-sided Fisher’s exact test, Padj < 10−5 and mutation ratio lower than 0.25) as intersections between all three of the genetic screens that were performed in this study. CCDC101 is also known as SGF29. c, HeLa cells, 293T cells, BJEH fibroblasts and N/TERT-1 keratinocytes were exposed to sgRNAs directed against the specified genes and pharmacologically stimulated for 9–12 h before immunoblotting (one representative experiment shown of n = 2 (HeLa and 293T) or n = 3 (BJEH and N/TERT-1) independent experiments). sgCTR, non-targeting control sgRNA. d, Clonal HAP1 knockout and stably reconstituted cells were treated as indicated (CCCP, tunicamycin and oligomycin (OM), 9 h; CDDO and GTPP, 11 h) and analysed by immunoblotting. e, HAP1 CHOPNeon cells of the indicated genotypes were treated for 9 h (CCCP, tunicamycin, CDDO) or 12 h (GTPP) and analysed by flow cytometry. Per genotype and treatment, the CHOPNeon signal was normalized to its DMSO control and statistical significance is indicated compared to identically treated wild-type cells (mean ± s.d. of n = 3 independent experiments; one-way ANOVA with Dunnett’s multiple comparisons correction). f, Wild-type, DELE1-deficient (clones #3 and #6) and stably reconstituted HAP1 CHOPNeon cells were transduced with lentiviral constructs that contain a non-targeting control sgRNA or an sgRNA directed against LONP1. CHOPNeon levels were analysed by flow cytometry. Per genotype, the CHOPNeon fluorescence detected in sgLONP1-treated cells is shown relative to the CHOPNeon fluorescence detected in the matching sgCTR-treated cells (n = 2 independent experiments). g, HeLa, 293T and HCT116 cells—either wild type or that were transiently exposed to an sgRNA directed against EIF2AK1 (HRI) or DELE1—were stimulated with CCCP for 1 h and the phosphorylation of eIF2α was analysed by immunoblotting (one representative experiment shown of two independent experiments).
Extended Data Fig. 6 DELE1 operates upstream of HRI, is cleaved and accumulates in the cytosol across different types of mitochondrial perturbation and different cellular systems.
a, b, HeLa wild-type, knockout and stably reconstituted cells were treated for 1 h (phosphorylated eIF2α) or 9 h (CHOP) with CCCP. Cells were analysed by immunoblotting (a) and the levels of phosphorylated eIF2α were quantified relative to ACTB (b; mean ± s.d. of n = 5 independent experiments; for each genotype, CCCP treatment was compared to DMSO using a paired two-tailed Student’s t-test). c, d, Subcellular localization of HA-tagged DELE1 or DELE1(∆MTS) in HeLa cells analysed by confocal microscopy. TRAP1 staining identifies mitochondria and nuclei were visualized with DAPI. e, Top, clonal DELE1-knockout HAP1 CHOPNeon cells were transiently transfected with full-length DELE1 or truncation mutants that lack the first 101 (DELE1(ΔMTS(N101))) or 115 (DELE1(ΔMTS(N115))) amino acids as indicated. Bottom, transfected cells were treated with CCCP or DMSO for 9 h and analysed by flow cytometry as in Fig. 2a (mean ± s.d. of n = 3 independent biological samples; two-way ANOVA with Tukey’s multiple comparisons correction; one representative experiment shown of three independent experiments). f, HeLa cells were transfected with DELE1-HA, treated with CCCP alone or CCCP and cycloheximide (CHX) as indicated and analysed for subcellular localization of DELE1. Mitochondria were stained with MitoTracker and nuclei were visualized using DAPI. g, CRISPR engineering of the DELE1 locus in 293T cells (introducing a C-terminal in-frame fusion with a triple HA tag), followed by treatment with CCCP alone or CCCP and cycloheximide and analysis by immunoblotting (one representative experiment shown of four independent experiments). h, Wild-type and HRI-knockout HAP1 cells were transfected with DELE1-HA and treated as indicated. The fate of the DELE1–HA protein was analysed by immunoblotting (n = 5 (HRI) or 2 (wild type) biologically independent clones). i, j, HCT116 (i) and HAP1 (j) cells were transfected with DELE1-HA, treated with the specified compounds and analysed for DELE1–HA cleavage and CHOP induction by immunoblotting (one representative experiment shown of two independent experiments). k, N/TERT-1 keratinocytes, hTERT+ (BJEH) fibroblasts and primary fibroblasts were transduced with DELE1–HA, stimulated for 4 h as indicated and assayed by immunoblotting. l, Wild-type 293T and HAP1 cells were transfected with DELE1-HA and treated with CCCP for 4 h. The localization of DELE1–HA was analysed as in Fig. 2d (one representative experiment shown of two independent experiments). m–o, Cells from k were stimulated as indicated for 4 h and the localization of DELE1–HA and MFN2 were analysed by confocal microscopy. TRAP1 staining (m), MitoTracker (n, o) and DAPI (o) were used to visualize cellular substructures (one representative experiment shown of two independent experiments). Scale bars, 10 μm (c, d, f, l–o).
Extended Data Fig. 7 Processing of full-length DELE1 into S-DELE1 is mediated by catalytically active OMA1.
a, Data from Fig. 1a, with statistical significance of mutation enrichment assessed and visualized in the same way (two-sided Fisher’s exact test, Padj < 0.05). Known mitochondrial proteases44,45 are highlighted in red. XRCC6BP1 is also known as ATP23. b, Wild-type HAP1 cells and clonal OMA1 knockout cells were transfected with DELE1-HA, treated with CCCP and assayed by immunoblotting. c, HeLa cells of the stated genotypes were transfected with DELE1-HA and pre-treated with the indicated protease inhibitors for 5 h, followed by the addition of CCCP for 2 h. The fates of DELE1, OMA1 and OPA1 were monitored by immunoblotting. DCI, 3,4-dichloroisocoumarin; pepstat. A, pepstatin A; o-phen, o-phenanthroline. d, Quantification of c. The fraction of S-DELE1 (versus L-DELE1) is indicated. Statistical significance was assessed using one-way ANOVA with Tukey’s multiple comparisons correction and represents the significance relative to the wild type plus CCCP condition (mean ± s.d. of n = 3 independent experiments). e, HeLa cells transiently exposed to an sgRNA directed against OMA1 were co-transfected with the indicated cDNAs and cleavage of DELE1–Flag–mNeon (FmN) after 2 h of CCCP treatment was monitored by immunoblotting (one representative experiment shown of two independent experiments). f, Workflow of the in vitro assay for DELE1 fate upon mitochondrial depolarization using mitochondria purified from wild-type or OMA1-deficient 293T cells that were transiently transfected with DELE1-HA. g, h, Purity analysis of isolated mitochondria using electron microscopy (g) and immunoblotting (h) (one representative experiment shown of two independent experiments for each). For immunoblotting, around 1% of the mitochondrial and 0.2% of the cytosolic fraction was used. The electron micrographs show a high proportion and purity of mitochondria, only slight contaminations of membrane fragments or other cell organelles and the complete absence of intact cells within the mitochondrial suspensions. Scale bars, 5 μm (left) or 1 μm (right). i–k, Isolated mitochondria from g, h were treated in vitro with FCCP and analysed for DELE1 processing. i, Mitochondria build up a membrane potential that remains stable for one hour (demonstrated by constant low fluorescence). FCCP (500 nM) was used to induce dissipation of the mitochondrial membrane potential, yielding an increase in fluorescence. j, After one hour of FCCP treatment, the mitochondria and supernatant were separated by centrifugation and the protein levels in both compartments were analysed by immunoblotting (top, long exposure; bottom, short exposure for DELE1–HA) k, The amount of S-DELE1 in the supernatants was quantified and compared to the wild type plus DMSO condition using two-way ANOVA with Tukey’s multiple comparisons correction. In i, k, data are mean ± s.d. of n = 5 independent experiments; j shows one representative experiment of n = 5 independent experiments, of which the purity analysis shown in g, h was performed for two independent experiments.
Extended Data Fig. 8 DELE1 cleavage and subcellular redistribution after CCCP treatment depend on OMA1 but not OPA1.
a–c, HeLa cells were treated with sgRNAs directed against OPA1, stimulated with CCCP and analysed as indicated together with wild-type HeLa cells by immunoblotting (a). OPA1 levels (b) and DELE1 cleavage (c) were quantified. Statistical significance was assessed using one-way ANOVA with Tukey’s multiple comparisons correction and represents the significance relative to the wild type (mean ± s.d. of n = 4 biologically independent samples). d, HAP1 cells were treated and analysed as in a (one representative experiment shown of two independent experiments). e, Data from Fig. 1a, with statistical significance of mutation enrichment assessed and visualized in the same way (two-sided Fisher’s exact test, Padj < 0.05). OPA1 is highlighted in red. f, HeLa cells were treated with an sgRNA directed against OPA1, stimulated with CCCP and analysed for DELE1 localization and OPA1 status, alongside wild-type and OMA1-knockout cells. Arrowheads highlight OPA1-deficient cells. One representative experiment shown of two independent experiments. Scale bars, 10 μm.
Extended Data Fig. 9 Physical interaction between DELE1 and HRI.
a, 293T cells were co-transfected with StrepTagII-HA-HRI and the indicated Flag-mNeon-tagged cDNAs before stimulation with CCCP as indicated. Cells were then lysed and StrepTagII-containing protein complexes were immobilized on Strep-Tactin beads and analysed by immunoblotting along with the input lysate. DELE1FL, full-length DELE1. b, A C-terminal in-frame fusion of a triple Flag tag was introduced into the endogenous EIF2AK1 (HRI) locus of 293T cells using CRISPR. c, Quantification of Fig. 3b. The bar chart shows the relative abundance of the slowly migrating species of HRI (phosphorylated HRI) compared to the faster migrating band. Data were normalized to the empty vector (black) and significance (compared to full-length DELE1 untreated, grey) was assessed using one-way ANOVA with Dunnett’s multiple comparisons correction (mean ± s.d. of n = 3 independent experiments). d, 293T cells were co-transfected with HRI-Flag and DELE1-HA and exposed to CCCP for the denoted amounts of time before immunoblotting. The shift in electrophoretic mobility of HRI upon activation is reversed by treatment of the lysates with alkaline phosphatase. e, 293T cells of the indicated genotypes were co-transfected with HRI-Flag and the specified HA-tagged constructs. After CCCP treatment, lysates were immobilized on anti-Flag beads and analysed by immunoblotting along with the input lysate (one representative experiment shown of four independent experiments). f, Quantification of e (mean ± s.d. of n = 4 independent experiments; two-way ANOVA with Tukey’s multiple comparisons correction). g, HeLa cells were transiently transfected with the indicated constructs, treated with CCCP or DMSO and analysed for subcellular localization of the transfected cDNAs by HA staining and confocal microscopy with cellular structures labelled as indicated. h, DELE1 mutants were transiently transfected into HeLa and 293T cells before exposure to CCCP for 2 h. The processing of DELE1–HA was monitored by immunoblotting. i, HeLa cells were transfected as indicated and analysed for subcellular localization as in g. j, 293T cells were co-transfected with HRI-Flag and the indicated StrepTagII-HA cDNAs. Cells were then lysed and StrepTagII-containing protein complexes were immobilized on Strep-Tactin beads and analysed by immunoblotting along with the input lysate. k, l, Clonal DELE1-knockout HAP1 cells were transiently transfected with the specified cDNAs together with mCherry and induction of CHOPNeon was monitored by flow cytometry as in Fig. 2a. Per genotype, data were normalized to the empty vector and statistical significance (compared to DELE1(∆MTS(N101))) was assessed using one-way ANOVA with Tukey’s multiple comparisons correction (mean ± s.d. of n = 4 independent experiments). m, Wild-type 293T cells were transiently transfected with HRI-Flag and the indicated HA-tagged cDNAs and processed as in e (one representative experiment shown of four independent experiments). n, DELE1-deficient HAP1 CHOPNeon cells were transiently transfected as indicated and analysed by flow cytometry 32 h after transfection. Per genotype, data were normalized to the empty vector (black) and statistical significance (compared to DELE(∆MTS)) was assessed using one-way ANOVA with Tukey’s multiple comparisons correction (mean ± s.d. of n = 3 independent experiments). o, Cell lysates from Fig. 3c were analysed by Coomassie staining before (lysate) and after exposure to GFP-TRAP beads (IP (GFP)). Scale bars, 10 μm (g, i).
Extended Data Fig. 10 Effects of haem and mitochondrial membrane potential on the OMA1–DELE1–HRI pathway.
a, Schematic depiction of haem metabolism. b, Data from Fig. 1a, with statistical significance of mutation enrichment assessed and visualized in the same way (two-sided Fisher’s exact test, Padj < 0.05). Haem-related genes from a are highlighted in red. c, Immunoprecipitation experiment as in Extended Data Fig. 9a in the presence of haemin during cell culture (20 μM for 12 h before lysis) and immunoprecipitation (8 μM in lysis and wash buffers). HMOX1 is induced in response to increased haem levels (one representative experiment shown of two independent experiments). d, Model of the pathway identified here. Mitochondrial stress activates OMA1, which leads to cleavage of DELE1 and its subsequent cytosolic binding to and activation of HRI, triggering the ISR. e, Wild-type HAP1 and HeLa cells were treated with CCCP, and the mitochondrial membrane potential was visualized by TMRM using a live-cell-imaging system. H2B-GFP was transfected to stain nuclei. Scale bars, 100 μm. f, g, HAP1 CHOPNeon cells were stimulated as indicated, and CHOPNeon levels (g) and the mitochondrial membrane potential (f) were quantified by flow cytometry. Statistical significance is depicted relative to DMSO-treated control cells (mean ± s.d. of n = 3 independent experiments; one-way ANOVA with Tukey’s multiple comparisons correction). h, HeLa cells were transfected with the indicated sgRNAs and the mitochondrial membrane potential was quantified after 1 h of CCCP treatment by flow cytometry. The TMRM signal was normalized to the non-targeting control sgRNA and DMSO control, and for CCCP-treated samples statistical significance was assessed relative to the non-targeting control sgRNA and CCCP (mean ± s.d. of n = 3 independent experiments; two-way ANOVA with Dunnett’s multiple comparisons correction). i, Volcano plots of RNA-seq analysis of the indicated samples. For every gene, the fold change in expression after CCCP treatment (versus DMSO treatment; x axis) is plotted against the P value (two-tailed Wald test in DESeq2; y axis). Genes with a significant differential expression (Padj < 10−12 (Benjamini–Hochberg)) are depicted in dark grey. ATF4 and CHOP target genes39,40 (green) and heat-shock proteins43 (red) from Fig. 3e are highlighted (n = 4 biologically independent samples per group). FAM129A is also known as NIBAN1.
Extended Data Fig. 11 Cellular fitness and mitochondrial function in cells in which the DELE1–HRI–eIF2α axis is perturbed.
a, HAP1 cells were infected with lentivirus expressing TagRFP and the indicated sgRNAs. A 1:1 mixture of infected and uninfected cells was treated for 48 h with 5 μM CCCP or DMSO and analysed by flow cytometry. The abundance of RFP+ sgRNA-containing cells after CCCP treatment was compared with the respective DMSO-treated sample bearing the same sgRNA and normalized to a non-targeting control sgRNA. Statistical significance was assessed compared to a second non-targeting control sgRNA using one-way ANOVA with Dunnett’s multiple comparisons correction (mean ± s.d. of n = 5 independent experiments). b, c, HAP1 cells of the indicated genotypes were treated as indicated for 48 h and cell survival was visualized using crystal violet (one representative experiment shown of three independent experiments). d–j, Wild-type and DELE1-knockout HeLa cells stably expressing the indicated cDNAs were subjected to different mitochondrial stressors and the oxygen consumption rate (OCR) was monitored. Within the same clonal background, no significant difference between DELE1-deficient and DELE1-proficient cells could be observed in non-mitochondrial oxygen consumption (e), basal respiration (f), maximal respiration (g), proton leak (h), ATP production (i) and spare respiratory capacity (j). Mean ± s.d. of n = 4 independent culture wells from one representative experiment of three independent experiments is shown; statistical significance was assessed using one-way ANOVA with Tukey’s multiple comparisons correction. k, l, Wild-type, MFN2-knockout and stably reconstituted HAP1 cells were lentivirally transduced with a construct encoding TagRFP together with a non-targeting control sgRNA or an sgRNA directed against EIF2AK1 (HRI) (k) or DELE1 (l) and the fraction of RFP+ cells was monitored over time. The abundance of sgDELE1 or sgHRI-containing cells was normalized to non-targeting sgRNA and to time 0 (mean ± s.d. of n = 3 biologically independent, separately infected and cultured wells per sgRNA and genotype; one-way ANOVA with Dunnett’s multiple comparisons correction; one representative experiment shown of two independent experiments). m, SH-SY5Y cells were exposed to the indicated sgRNAs and treated as in Extended Data Fig. 5c (one representative experiment shown of four independent experiments). n, o, Processing and cellular localization of DELE1–HA in SH-SY5Y cells, as in Extended Data Fig. 6k, o. MFN2–Flag–mNeon served as a control (one representative experiment shown of two independent experiments). Scale bars, 10 μm.
Supplementary information
41586_2020_2076_MOESM1_ESM.pdf
Supplementary Figures Supplementary Figure 1: Uncropped blot source data. All antibodies for one panel were probed on the same membrane, except for Ext. Data Fig. 1a where HSPD1 and ACTB were probed on the same membrane and CHOP on a different membrane and for Fig. 3c where GST and GFP were probed on two different membranes. Cropping to achieve the final figure panel was performed in Adobe Illustrator or Photoshop.
41586_2020_2076_MOESM3_ESM.xlsx
Supplementary Tables Supplementary Tables 1-3: Results of haploid genetic screens for CHOPNeon regulators using CCCP (Supplementary Table 1), tunicamycin (Supplementary Table 2) and CDDO (Supplementary Table 3). Data were processed as described in the methods section. The columns are: gene symbol, mutations identified in the respective gene in CHOPNeon-low cells, all other mutations identified in CHOPNeon-low cells, mutations identified in the respective gene in CHOPNeon-high cells, all other mutations identified in CHOPNeon-high cells, P-value, FDR-corrected P-value9. n = 2.12 × 107 (Supplementary Table 1), 2.07 × 107 (Supplementary Table 2), or 1.90 × 107 (Supplementary Table 3) interrogated single cells.
Supplementary Tables
Supplementary Table 4: Oligonucleotides used in this study.
41586_2020_2076_MOESM5_ESM.xlsx
Supplementary Tables Supplementary Table 5: Antibodies used in this study. WB, Western Blot; FACS, fluorescence-activated cell sorting; IF, Immunofluorescence; IP, Immunoprecipitation.
41586_2020_2076_MOESM6_ESM.xlsx
Supplementary Tables Supplementary Tables 6-10: Results of RNAseq analysis upon CCCP treatment: HAP1 wild-type cells (Supplementary Table 6), wild-type cells co-treated with ISRIB (Supplementary Table 7), HRI knockout cells (Supplementary Table 8), DELE1 knockout cells (Supplementary Table 9), and DELE1 knockout cells stably reconstituted with DELE1 (Supplementary Table 10). Data were processed as described in the methods section (n=4 biologically independent samples per group). The columns are: gene symbol, mean of normalized counts for all samples (baseMean), log2 fold change (MLE): condition treated (CCCP) vs. untreated (DMSO), standard error: condition treated vs untreated, Wald statistic: condition treated vs untreated, Wald test P-value (two-tailed): condition treated vs. untreated, Benjamini-Hochberg adjusted P-value.
Source data
Rights and permissions
About this article
Cite this article
Fessler, E., Eckl, EM., Schmitt, S. et al. A pathway coordinated by DELE1 relays mitochondrial stress to the cytosol. Nature 579, 433–437 (2020). https://doi.org/10.1038/s41586-020-2076-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2076-4
This article is cited by
-
Stress response silencing by an E3 ligase mutated in neurodegeneration
Nature (2024)
-
Mammalian integrated stress responses in stressed organelles and their functions
Acta Pharmacologica Sinica (2024)
-
Cytosolic retention of HtrA2 during mitochondrial protein import stress triggers the DELE1-HRI pathway
Communications Biology (2024)
-
DELE1 haploinsufficiency causes resistance to mitochondrial stress-induced apoptosis in monosomy 5/del(5q) AML
Leukemia (2024)
-
NAD+ dependent UPRmt activation underlies intestinal aging caused by mitochondrial DNA mutations
Nature Communications (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.