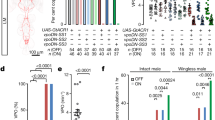
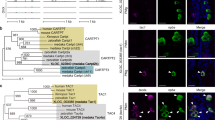
Abstract
Mating and egg laying are tightly cooordinated events in the reproductive life of all oviparous females. Oviposition is typically rare in virgin females but is initiated after copulation. Here we identify the neural circuitry that links egg laying to mating status in Drosophila melanogaster. Activation of female-specific oviposition descending neurons (oviDNs) is necessary and sufficient for egg laying, and is equally potent in virgin and mated females. After mating, sex peptide—a protein from the male seminal fluid—triggers many behavioural and physiological changes in the female, including the onset of egg laying1. Sex peptide is detected by sensory neurons in the uterus2,3,4, and silences these neurons and their postsynaptic ascending neurons in the abdominal ganglion5. We show that these abdominal ganglion neurons directly activate the female-specific pC1 neurons. GABAergic (γ-aminobutyric-acid-releasing) oviposition inhibitory neurons (oviINs) mediate feed-forward inhibition from pC1 neurons to both oviDNs and their major excitatory input, the oviposition excitatory neurons (oviENs). By attenuating the abdominal ganglion inputs to pC1 neurons and oviINs, sex peptide disinhibits oviDNs to enable egg laying after mating. This circuitry thus coordinates the two key events in female reproduction: mating and egg laying.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The datasets generated during the current study are available from the corresponding author on reasonable request.
References
Kubli, E. The sex-peptide. BioEssays14, 779–784 (1992).
Yapici, N., Kim, Y. J., Ribeiro, C. & Dickson, B. J. A receptor that mediates the post-mating switch in Drosophila reproductive behaviour. Nature451, 33–37 (2008).
Häsemeyer, M., Yapici, N., Heberlein, U. & Dickson, B. J. Sensory neurons in the Drosophila genital tract regulate female reproductive behavior. Neuron61, 511–518 (2009).
Yang, C. H. et al. Control of the postmating behavioral switch in Drosophila females by internal sensory neurons. Neuron61, 519–526 (2009).
Feng, K., Palfreyman, M. T., Häsemeyer, M., Talsma, A. & Dickson, B. J. Ascending SAG neurons control sexual receptivity of Drosophila females. Neuron83, 135–148 (2014).
Auer, T. O. & Benton, R. Sexual circuitry in Drosophila. Curr. Opin. Neurobiol. 38, 18–26 (2016).
Kvitsiani, D. & Dickson, B. J. Shared neural circuitry for female and male sexual behaviours in Drosophila. Curr. Biol. 16, R355–R356 (2006).
Demir, E. & Dickson, B. J. fruitless splicing specifies male courtship behavior in Drosophila. Cell121, 785–794 (2005).
Kupfermann, I. & Weiss, K. R. The command neuron concept. Behav. Brain Sci. 1, 3–10 (1978).
Luan, H., Peabody, N. C., Vinson, C. R. & White, B. H. Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron52, 425–436 (2006).
Dionne, H., Hibbard, K. L., Cavallaro, A., Kao, J. C. & Rubin, G. M. Genetic reagents for making split-GAL4 lines in Drosophila. Genetics209, 31–35 (2018).
Tirian, L. & Dickson, B. J. The VT GAL4, LexA, and split-GAL4 driver line collections for targeted expression in the Drosophila nervous system. Preprint at bioRxiv https://doi.org/10.1101/198648 (2017).
Klapoetke, N. C. et al. Independent optical excitation of distinct neural populations. Nat. Methods11, 338–346 (2014).
Nern, A., Pfeiffer, B. D. & Rubin, G. M. Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proc. Natl Acad. Sci. USA112, E2967–E2976 (2015).
Zheng, Z. et al. A complete electron microscopy volume of the brain of adult Drosophila melanogaster. Cell174, 730–743 (2018).
Yang, C. H., Belawat, P., Hafen, E., Jan, L. Y. & Jan, Y. N. Drosophila egg-laying site selection as a system to study simple decision-making processes. Science319, 1679–1683 (2008).
Kimura, K., Sato, C., Koganezawa, M. & Yamamoto, D. Drosophila ovipositor extension in mating behavior and egg deposition involves distinct sets of brain interneurons. PLoS ONE 10, e0126445 (2015).
McKellar, C. E. et al. Threshold-based ordering of sequential actions during Drosophila courtship. Curr. Biol. 29, 426–434 (2019).
Zhou, C., Pan, Y., Robinett, C. C., Meissner, G. W. & Baker, B. S. Central brain neurons expressing doublesex regulate female receptivity in Drosophila. Neuron83, 149–163 (2014).
Cury, K. M., Prud’homme, B. & Gompel, N. A short guide to insect oviposition: when, where and how to lay an egg. J. Neurogenet. 33, 75–89 (2019).
Gou, B., Zhu, E., He, R., Stern, U. & Yang, C. H. High throughput assay to examine egg-laying preferences of individual Drosophila melanogaster. J. Vis. Exp. 109, e53716 (2016).
Thomas, A. Nervous control of egg progression into the common oviduct and genital chamber of the stick-insect Carausius morosus. J. Insect Physiol. 25, 811–823 (1979).
Bath, D. E. et al. FlyMAD: rapid thermogenetic control of neuronal activity in freely walking Drosophila. Nat. Methods11, 756–762 (2014).
Inagaki, H. K. et al. Optogenetic control of Drosophila using a red-shifted channelrhodopsin reveals experience-dependent influences on courtship. Nat. Methods11, 325–332 (2014).
von Philipsborn, A. C. et al. Neuronal control of Drosophila courtship song. Neuron69, 509–522 (2011).
Ribeiro, I. M. A. et al. Visual projection neurons mediating directed courtship in Drosophila. Cell174, 607–621 (2018).
Backhaus, B., Sulkowski, E. & Schlote, F. W. A semi-synthetic, general-purpose medium for Drosophila melanogaster. Drosoph. Inf. Serv. 60, 210–212 (1984).
Jenett, A. et al. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2, 991–1001 (2012).
Shirangi, T. R., Wong, A. M., Truman, J. W. & Stern, D. L. Doublesex regulates the connectivity of a neural circuit controlling Drosophila male courtship song. Dev. Cell37, 533–544 (2016).
Otsuna, H., Ito, M. & Kawase, T. Color depth MIP mask search: a new tool to expedite Split-GAL4 creation. Preprint at bioRxiv https://doi.org/10.1101/318006 (2018).
Schneider-Mizell, C. M. et al. Quantitative neuroanatomy for connectomics in Drosophila. eLife5, e12059 (2016).
Wilson, R. I. & Laurent, G. Role of GABAergic inhibition in shaping odor-evoked spatiotemporal patterns in the Drosophila antennal lobe. J. Neurosci. 25, 9069–9079 (2005).
Chen, T. W. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature499, 295–300 (2013).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods9, 676–682 (2012).
Thévenaz, P., Ruttimann, U. E. & Unser, M. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 7, 27–41 (1998).
Wu, M. et al. Visual projection neurons in the Drosophila lobula link feature detection to distinct behavioral programs. eLife5, e21022 (2016).
Meissner, G. W. et al. Mapping neurotransmitter identity in the whole-mount Drosophila brain using multiplex high-throughput fluorescence in situ hybridization. Genetics211, 473–482 (2019).
von Reyn, C. R. et al. A spike-timing mechanism for action selection. Nat. Neurosci. 17, 962–970 (2014).
Acknowledgements
We thank the Janelia FlyLight, Fly Facility, Project Technical Resources, Molecular Biology, Functional Connectome and Experimental Technology teams for technical assistance; A. Edmonson-Stait and G. Jefferis for initial tracing of one of the oviIN cells; U. Heberlein, K. Feng and V. Vijayan for comments on the manuscript; and V. Vijayan and G. Maimon for sharing preliminary oviDN calcium-imaging data. This work was funded by the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
B.J.D., F.W. and K.W. conceived the study and wrote the manuscript. F.W. and K.W. performed all experiments and analysed the data. N.F., C.P., T.Y., F.W. and R.P. reconstructed selected neurons and synapses in the FAFB electron microscopy volume, which was provided before publication by D.B.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review informationNature thanks Rebecca Yang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Split-GAL4 driver lines targeting oviDNs, SPSNs, SAG neurons, pC1 neurons, oviENs and oviINs.
Confocal images of the central nervous system from female and male flies carrying the indicated split-GAL4 driver lines as well as UAS-myrFLAG or UAS-CsChrimson-mVenus. Samples were stained with the monoclonal antibody nc82 to reveal all synapses (magenta), and with anti-FLAG or anti-GFP to reveal the membranes of targeted neurons (green). Scale bar, 100 μm. Both oviDN-SS1 and oviDN-SS2 label a single oviDNa and a single oviDNb cell in each hemisphere; oviDN-SS2 also weakly labels an unrelated cell (pMP1) that is present in both sexes.
Extended Data Fig. 2 Expression of fru and dsx in oviDNs and pC1 neurons.
Confocal images of female brains showing the co-labelling of oviDN-SS lines with fru-LexA but not dsx-LexA, and of the pC1-SS1 line with dsx-LexA but not fru-LexA. Scale bars, 20 μm.
Extended Data Fig. 3 Neurotransmitter types revealed by fluorescence in situ hybridization.
Confocal images showing the expression of GAD1, ChAT and vGluT in oviDNs, SAG neurons, pC1 neurons, oviENs and oviINs in female brains. Red arrows indicate cell bodies of interest. Scale bars, 20 μm.
Extended Data Fig. 4 oviDNs are required for oviposition but not copulation.
a, Number of GFP-expressing neurons in female brains of the indicated genotypes. **P < 0.01 by Wilcoxon test. b, Total number of eggs laid by individual mated females over 10 consecutive days after mating, shown as mean ± s.e.m. Note the values of zero for both oviDN-ablated genotypes at all time points. ***P < 0.001 by Wilcoxon test. c, Cumulative traces showing the percentage of females copulating over a 30-min observation period.
Extended Data Fig. 5 Stochastic labelling and unsilencing of oviDNs.
a, Images of two female samples in which a single oviDNa or oviDNb cell is labelled, as shown in Fig. 1b. Arrowheads indicate branches that are present in oviDNb (solid) but absent in oviDNa (open). The branch that is labelled by arrowhead 1 was primarily used to distinguish oviDNa from oviDNb. b, Example images of brains in which oviDNs were either silenced (red; Kir2.1::tdTomato) or unsilenced (green; mCD8::GFP). The number of unsilenced oviDNs in each sample is shown. Green arrowheads indicate distinctive branches of oviDNb. Brains were counterstained with nc82 (blue). Scale bar, 100 μm. c, Number of eggs laid in the five days after mating by mated females with different oviDNs unsilenced. ***P < 0.001, **P < 0.01 by Wilcoxon test. Scatter plots show mean ± s.e.m. d, Confocal images of two samples in which a single oviDN was loaded with neurobiotin during whole-cell recording. The samples were stained with streptavidin (to reveal the recorded cell, yellow) and nc82 (blue). Arrowheads indicate oviDNb-specific branches. Scale bars, 100 μm.
Extended Data Fig. 6 Sequence of oviposition actions after oviDN stimulation.
Example ethograms showing the onsets of oviposition actions in mated females after photoactivation (3 s) of oviDNs at varying light intensities. Each row represents a single female.
Extended Data Fig. 7 Anatomical and functional characterization of pC1 neurons.
a. Confocal images of single pC1 neurons in the female brain, as shown in Fig. 3d. Arrowheads indicate the presence (solid) or absence (open) of subtype-specific branches. b, Confocal images of neurobiotin-filled pC1 neurons from which whole-cell patch recordings were obtained, indicating the branches that were used for subtype identification as in a. c, d, Number of eggs laid by virgin females during a one-hour (c) or three-day (d) period in which either SAG or pC1 neurons were optogenetically silenced. e, Basal GCaMP6s signals in pC1 cell bodies in virgin and mated females. ***P < 0.001 by Wilcoxon test; scatter plots show mean ± s.e.m. (d, e).
Extended Data Fig. 8 Egg-laying substrate preferences and substrate-evoked calcium responses in oviDNs, oviENs and oviINs.
a, Image of the egg-laying chambers in each of which an individual mated female had laid numerous eggs. Chambers with plain agarose (blue box), agarose containing 150 mM sucrose (red box) and plastic surface (green box) are indicated. b, Total number of eggs laid by individual mated females in a 12-h observation period. **P < 0.01, ***P < 0.001 by Wilcoxon test. c, Preference indices showing the preference of female flies for laying eggs on different substrates. Preference index (PI) is calculated as (number of eggs on plain agarose – number of eggs on other substrate)/total number of eggs. Data are mean ± s.e.m. d, Projected images of oviDNs (top), oviENs (middle) and oviINs (bottom) expressing GCaMP6s, showing ROIs for quantification. e, Example ΔF/F0 traces for each ROI upon presentation of the indicated substrates, in virgin (left) and mated (right) females. Horizontal bars indicate presentation of the substrate. Darker traces are averaged from six trials (lighter traces).
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1 and 2.
Video 1
: Optogenetic activation of oviDNs elicits oviposition behaviour. A montage of video clips of 16 mated oviDN-SS2 UAS-Chrimson females upon 5 s of continuous 625 nm illumination at 200 µW/mm2), shown at half speed (100 fps).
Video 2
: oviDNs reconstructed in female brain EM volume. Two oviDNa cells (blue and green) and one oviDNb cell (red) reconstructed in the right hemisphere of the FAFB EM volume.
Video 3
: pC1 neurons reconstructed in female brain EM volume. Five pC1 neurons reconstructed in the right hemisphere of the FAFB EM volume. pC1a, pC1c, and pC1e were fully traced; pC1b and pC1d were only partially reconstructed.
Video 4
: SAG neurons are presynaptic to pC1a neurons. A pair of SAG neurons (blue and green) partially reconstructed in the FAFB EM volume, and one fully-traced pC1a neuron (red) in the right hemisphere. The location of synapses between SAG and pC1a neurons are indicated by yellow or cyan balls.
Video 5
: oviEN and oviIN are presynaptic to oviDNs. oviDN (blue), oviEN (pink), oviIN (green) and pC1neurons (red) reconstructed in the FAFB EM volume in the right hemisphere.
Rights and permissions
About this article
Cite this article
Wang, F., Wang, K., Forknall, N. et al. Neural circuitry linking mating and egg laying in Drosophila females. Nature 579, 101–105 (2020). https://doi.org/10.1038/s41586-020-2055-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2055-9
This article is cited by
-
The spatial and temporal structure of neural activity across the fly brain
Nature Communications (2023)
-
Odor-regulated oviposition behavior in an ecological specialist
Nature Communications (2023)
-
A rise-to-threshold process for a relative-value decision
Nature (2023)
-
Flexible neural control of transition points within the egg-laying behavioral sequence in Drosophila
Nature Neuroscience (2023)
-
Neuronal substrates of egg-laying behaviour at the abdominal ganglion of Drosophila melanogaster
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.