Abstract
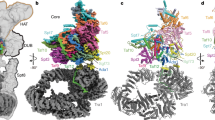
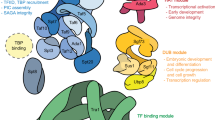
SAGA (Spt–Ada–Gcn5–acetyltransferase) is a 19-subunit complex that stimulates transcription via two chromatin-modifying enzymatic modules and by delivering the TATA box binding protein (TBP) to nucleate the pre-initiation complex on DNA, a pivotal event in the expression of protein-encoding genes1. Here we present the structure of yeast SAGA with bound TBP. The core of the complex is resolved at 3.5 Å resolution (0.143 Fourier shell correlation). The structure reveals the intricate network of interactions that coordinate the different functional domains of SAGA and resolves an octamer of histone-fold domains at the core of SAGA. This deformed octamer deviates considerably from the symmetrical analogue in the nucleosome and is precisely tuned to establish a peripheral site for TBP, where steric hindrance represses binding of spurious DNA. Complementary biochemical analysis points to a mechanism for TBP delivery and release from SAGA that requires transcription factor IIA and whose efficiency correlates with the affinity of DNA to TBP. We provide the foundations for understanding the specific delivery of TBP to gene promoters and the multiple roles of SAGA in regulating gene expression.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB) under accession codes EMD-10438 (SAGA–TBP), EMD-10440 (SAGA–TBP, refined for Tra1 lobe), EMD-10441 (SAGA–TBP, refined for main lobe), EMD-10446 (SAGA–TBP, low-pass-filtered), EMD-10448 (SAGA–TBP, focused classification on TBP), EMD-10447 (SAGA without TBP). The model coordinates for SAGA–TBP were deposited in the Protein Data Bank (PDB) under the accession codes 6TB4 and 6TBM (including Spt8 and DUB).
References
Helmlinger, D. & Tora, L. Sharing the SAGA. Trends Biochem. Sci. 42, 850–861 (2017).
Roeder, R. G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 21, 327–335 (1996).
Sainsbury, S., Bernecky, C. & Cramer, P. Structural basis of transcription initiation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 16, 129–143 (2015).
Hahn, S. & Young, E. T. Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics 189, 705–736 (2011).
Larschan, E. & Winston, F. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 15, 1946–1956 (2001).
Bhaumik, S. R. & Green, M. R. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol. Cell. Biol. 22, 7365–7371 (2002).
Baptista, T. et al. SAGA is a general cofactor for RNA polymerase II transcription. Mol. Cell 68, 130–143 (2017).
Warfield, L. et al. Transcription of nearly all yeast RNA polymerase II-transcribed genes is dependent on transcription factor TFIID. Mol. Cell. 68, 118–129 (2017).
Lee, K. K. et al. Combinatorial depletion analysis to assemble the network architecture of the SAGA and ADA chromatin remodeling complexes. Mol. Syst. Biol. 7, 503 (2011).
Köhler, A., Zimmerman, E., Schneider, M., Hurt, E. & Zheng, N. Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module. Cell 141, 606–617 (2010).
Samara, N. L. et al. Structural insights into the assembly and function of the SAGA deubiquitinating module. Science 328, 1025–1029 (2010).
Sun, J. et al. Structural basis for activation of SAGA histone acetyltransferase Gcn5 by partner subunit Ada2. Proc. Natl Acad. Sci. USA 115, 10010–10015 (2018).
Díaz-Santín, L. M., Lukoyanova, N., Aciyan, E. & Cheung, A. C. Cryo-EM structure of the SAGA and NuA4 coactivator subunit Tra1 at 3.7 angstrom resolution. eLife 6, e28384 (2017).
Sharov, G. et al. Structure of the transcription activator target Tra1 within the chromatin modifying complex SAGA. Nat. Commun. 8, 1556 (2017).
Setiaputra, D. et al. Conformational flexibility and subunit arrangement of the modular yeast Spt–Ada–Gcn5 acetyltransferase complex. J. Biol. Chem. 290, 10057–10070 (2015).
Wu, P. Y., Ruhlmann, C., Winston, F. & Schultz, P. Molecular architecture of the S. cerevisiae SAGA complex. Mol. Cell 15, 199–208 (2004).
Gangloff, Y. G., Romier, C., Thuault, S., Werten, S. & Davidson, I. The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem. Sci. 26, 250–257 (2001).
Grant, P. A. et al. A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 94, 45–53 (1998).
Kolesnikova, O. et al. Molecular structure of promoter-bound yeast TFIID. Nat. Commun. 9, 4666 (2018).
Patel, A. B. et al. Structure of human TFIID and mechanism of TBP loading onto promoter DNA. Science 362, eaau8872 (2018).
Eisenmann, D. M., Arndt, K. M., Ricupero, S. L., Rooney, J. W. & Winston, F. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 6, 1319–1331 (1992).
Mohibullah, N. & Hahn, S. Site-specific cross-linking of TBP in vivo and in vitro reveals a direct functional interaction with the SAGA subunit Spt3. Genes Dev. 22, 2994–3006 (2008).
Sermwittayawong, D. & Tan, S. SAGA binds TBP via its Spt8 subunit in competition with DNA: implications for TBP recruitment. EMBO J. 25, 3791–3800 (2006).
Birck, C. et al. Human TAFII28 and TAFII18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family. Cell 94, 239–249 (1998).
Hoffmann, A. et al. A histone octamer-like structure within TFIID. Nature 380, 356–359 (1996).
Luger, K., Mäder, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 (1997).
Nikolov, D. B. et al. Crystal structure of a TFIIB–TBP–TATA-element ternary complex. Nature 377, 119–128 (1995).
Wu, P. Y. & Winston, F. Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex. Mol. Cell. Biol. 22, 5367–5379 (2002).
Geiger, J. H., Hahn, S., Lee, S. & Sigler, P. B. Crystal structure of the yeast TFIIA/TBP/DNA complex. Science 272, 830–836 (1996).
Tan, S., Hunziker, Y., Sargent, D. F. & Richmond, T. J. Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature 381, 127–134 (1996).
Wollmann, P. et al. Structure and mechanism of the Swi2/Snf2 remodeller Mot1 in complex with its substrate TBP. Nature 475, 403–407 (2011).
Imbalzano, A. N., Zaret, K. S. & Kingston, R. E. Transcription factor (TF) IIB and TFIIA can independently increase the affinity of the TATA-binding protein for DNA. J. Biol. Chem. 269, 8280–8286 (1994).
Warfield, L., Ranish, J. A. & Hahn, S. Positive and negative functions of the SAGA complex mediated through interaction of Spt8 with TBP and the N-terminal domain of TFIIA. Genes Dev. 18, 1022–1034 (2004).
Petrenko, N., Jin, Y., Dong, L., Wong, K. H. & Struhl, K. Requirements for RNA polymerase II preinitiation complex formation in vivo. eLife 8, e43654 (2019).
Anandapadamanaban, M. et al. High-resolution structure of TBP with TAF1 reveals anchoring patterns in transcriptional regulation. Nat. Struct. Mol. Biol. 20, 1008–1014 (2013).
Han, Y., Luo, J., Ranish, J. & Hahn, S. Architecture of the Saccharomyces cerevisiae SAGA transcription coactivator complex. EMBO J. 33, 2534–2546 (2014).
Kamata, K. et al. C-terminus of the Sgf73 subunit of SAGA and SLIK is important for retention in the larger complex and for heterochromatin boundary function. Genes Cells 18, 823–837 (2013).
Elias-Villalobos, E., Toullec, D., Faux, C., Séveno, M. & Helmlinger, D. Chaperone-mediated ordered assembly of the SAGA and NuA4 transcription co-activator complexes. Nat. Commun. 10, 5237 (2019).
Saint, M. et al. The TAF9 C-terminal conserved region domain is required for SAGA and TFIID promoter occupancy to promote transcriptional activation. Mol. Cell. Biol. 34, 1547–1563 (2014).
Huisinga, K. L. & Pugh, B. F. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 13, 573–585 (2004).
Ravarani, C. N., Chalancon, G., Breker, M., de Groot, N. S. & Babu, M. M. Affinity and competition for TBP are molecular determinants of gene expression noise. Nat. Commun. 7, 10417 (2016).
Gupta, K. et al. Architecture of TAF11/TAF13/TBP complex suggests novel regulation properties of general transcription factor TFIID. eLife 6, e30395 (2017).
Ranish, J. A., Lane, W. S. & Hahn, S. Isolation of two genes that encode subunits of the yeast transcription factor IIA. Science 255, 1127–1129 (1992).
Mittal, C., Culbertson, S. J. & Shogren-Knaak, M. A. Distinct requirements of linker DNA and transcriptional activators in promoting SAGA-mediated nucleosome acetylation. J. Biol. Chem. 293, 13736–13749 (2018).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Grant, T., Rohou, A. & Grigorieff, N. cisTEM, user-friendly software for single-particle image processing. eLife 7, e35383 (2018).
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014).
van Heel, M., Harauz, G., Orlova, E. V., Schmidt, R. & Schatz, M. A new generation of the IMAGIC image processing system. J. Struct. Biol. 116, 17–24 (1996).
Madeira, F. et al. The EMBL–EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641 (2019).
Yang, J. & Zhang, Y. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 43, W174-81 (2015).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Källberg, M. et al. Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 7, 1511–1522 (2012).
Buchan, D. W., Minneci, F., Nugent, T. C., Bryson, K. & Jones, D. T. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 41, W349-57 (2013).
Terwilliger, T. C. Rapid model building of alpha-helices in electron-density maps. Acta Crystallogr. D 66, 268–275 (2010).
Goddard, T. D., Huang, C. C. & Ferrin, T. E. Visualizing density maps with UCSF Chimera. J. Struct. Biol. 157, 281–287 (2007).
Goddard, T. D. et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Zhou, Q. & Berk, A. J. The yeast TATA-binding protein (TBP) core domain assembles with human TBP-associated factors into a functional TFIID complex. Mol. Cell. Biol. 15, 534–539 (1995).
Kelleher, R. J., III et al. Yeast and human TFIIDs are interchangeable for the response to acidic transcriptional activators in vitro. Genes Dev. 6, 296–303 (1992).
Ozer, J., Lezina, L. E., Ewing, J., Audi, S. & Lieberman, P. M. Association of transcription factor IIA with TATA binding protein is required for transcriptional activation of a subset of promoters and cell cycle progression in Saccharomyces cerevisiae. Mol. Cell. Biol. 18, 2559–2570 (1998).
Laprade, L., Rose, D. & Winston, F. Characterization of new Spt3 and TATA-binding protein mutants of Saccharomyces cerevisiae: Spt3 TBP allele-specific interactions and bypass of Spt8. Genetics 177, 2007–2017 (2007).
Acknowledgements
We thank D. Devys and L. Tora for carefully reading the manuscript and for advice; R. Wagner and B. Séraphin for advice during the initial stages of this project; and R. Ben-Shem for graphics expertise. We acknowledge support from the Institut National de la Santé et de la Recherche Médicale (INSERM), the Centre National pour la Recherche Scientifique (CNRS), the Association pour la Recherche sur le Cancer (ARC), the Ligue contre le Cancer, ANR-15-CE11-0022-01 and ANR-10-LABX-0030-INRT, a French State fund managed by the Agence Nationale de la Recherche under the frame program Investissements d’Avenir ANR-10- IDEX-0002-02. We acknowledge the use of resources of the French Infrastructure for Integrated Structural Biology FRISBI ANR-10-INBS-05 and of Instruct-ERIC.
Author information
Authors and Affiliations
Contributions
P.S. and A.B.-S. designed the study; A.B.-S. designed the SAGA purification method and reconstituted the SAGA–TBP complex; A.B.-S. and C.C. defined conditions for grid preparation and freezing; C.C. and G.P. froze grids; G.P. collected and analysed cryo-EM data; A.F., G.P. and A.B.-S. interpreted the maps by fitting crystal coordinates and model building; O.K. purified TBP, TFIIA and SAGA and used them to perform pull-down and gel-shift assays; P.S. and A.B.-S. supervised the work; G.P. and O.K. prepared figures; and A.B.-S. and P.S. wrote the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Steve Hahn and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Subunit and domain organization of the yeast SAGA complex.
a, SAGA subunits are organized into the Tra1 lobe and the main lobe. The main lobe is divided into enzymatic HAT and DUB modules, a central module and a TBP-binding module. Overlaps appear as the central module is a scaffold for the assembly of all other modules. Abbreviations used owing to space limitations: Taf5 lissencephaly homology (LisH) domain (T5L), Taf6 linker domain (N83–S218, T6L), Taf9 C tail (N110–L139, T9T), Sgf73 NTD (M1–K69, ND), Sgf73 linker region (I70–H234, S73L), Sgf73 anchoring domain (E235–L277, S73A), Spt20 HIT domain interacting with Tra1 (M325–L340, HIT), Ada3 anchoring domains (K398–L471, A3A1) and (Y509–E557, A3A2) and bromodomain (BRD). The SEP domain found in Spt20 is named after S. cerevisiae Shp1, Drosophila melanogaster eyes closed gene (Eyc) and vertebrate P47. The FAT repeats are divided into three TRD domains and one HEAT-repeats domain (HRD). We defined three bridge-forming domains involved in connecting Tra1 to the central module: the Spt20 bridge (K184–K336, S20B), the Taf12 bridge (T430–K502, T12B) and the Spt3 bridge (E268–I304, S3B). The histone-fold domains, present in seven subunits, are highlighted as yellow boxes. b, Schematic representation and helix nomenclature for histone-fold domains. c, Original micrograph of frozen-hydrated P. pastoris SAGA–TBP complex. d, Two-dimensional class averages showing high-resolution structural features.
Extended Data Fig. 2 Cryo-EM data-analysis strategy and resolution assessment of the cryo-EM structures.
a, Image-processing strategy used to obtain the 3.5 Å resolution maps of the SAGA–TBP complex. Maps coloured in rainbow represent the local resolution of the reconstructions. FSC curves are depicted as a function of resolution in angstrom for the entire SAGA–TBP complex (black), the Tra1 lobe (blue) and the main lobe (red). cryoSPARC v.2 and cisTEM were used to generate the 3.5 Å resolution maps of the Tra1 and main lobes. RELION 3 was used for masked refinements and classifications of flexible regions, corresponding to TBP, the DUB module and Spt8. Dotted circles show the regions that were used to dock the crystal structures of TBP (PDB: 1TBP), Spt8 (homology model by Swiss-Model using PDB 4J87 as template) and the DUB module (PDB: 6AQR). b, c, Representative regions illustrating the quality of the cryo-EM map and the high-resolution structural features. Cryo-EM map and atomic model showing that side chains are clearly identified. b, Histone-fold domain of Taf9. c, Part of the Taf5 WD40 domain.
Extended Data Fig. 3 Fitting of TBP and Spt8 into the cryo-EM map.
a, Pairwise alignment of TBP from P. pastoris and S. cerevisiae. Eight residues (out of 180) differ between the two organisms within the conserved C-terminal part of TBP (marked by a dotted line). These residues are mapped on the structure of cTBP (PDB: 1YTF). None of these residues occur in regions that contact SAGA (highlighted in red). b, Pairwise alignment of TFIIA subunits, Toa1 and Toa2, from P. pastoris and S. cerevisiae. The evolutionary conserved and structured domains of TFIIA (marked by dotted lines) show 60% identity and 80% similarity between the two organisms. It is worth noting that TBP and TFIIA have both been shown to be highly similar even across large evolutionary distances. For example, TBP, as well as Toa2, from yeast and human are functionally interchangeable in Pol II transcription61,62,63 and yeast TFIIA complements a mammalian in vitro transcription system depleted of TFIIA43. Hence, TBP and TFIIA from S. cerevisiae, which are easier to overproduce in E. coli, are valid substitutes in our experimental system for their homologues from the closely related budding yeast P. pastoris. c, Cryo-EM reconstructions determined in the presence (green) and in the absence (pink) of TBP. The enlarged panel shows the superimposition of both maps and highlights the additional density corresponding to TBP. d, Fitting of the cTBP crystal structure into the additional density observed in the SAGA–TBP complex. The α-loop denotes an α-helix from the linker connecting the two histone folds in Spt3. e, f, Fitting of the WD40 repeat of subunit Spt8 into the cryo-EM map of the SAGA–TBP complex next to TBP.
Extended Data Fig. 4 Comparing the arrangement of common SAGA and TFIID subunits.
a, The Taf5 NTD and Taf6 HEAT repeats have different positions in SAGA and in TFIID. The structures of Taf5 NTD, Taf6 HEAT repeats and Taf5 WD40 domain in SAGA (left) and in the twin lobe of TFIID19 (right) are depicted. For the sake of clarity, all other parts of SAGA and TFIID were removed. To compare the position of the Taf5 NTD and Taf6 HEAT repeats between SAGA and TFIID, both complexes were aligned to have the same orientation of the Taf5 WD40 domain. The top face of the WD40 repeat, which is to a large extent free in TFIID, is nearly completely engaged in SAGA. The positions of the Taf5 NTD and Taf6 HEAT repeats are defined by direct interactions of the two domains between themselves, with the WD40 surface, and with the α-helix A352–D364 of Taf5. b, SAGA and TFIID share five subunits (Taf5 and the four histone fold-containing subunits Taf6, Taf9, Taf10 and Taf12). These subunits are present as one and two copies in SAGA and TFIID, respectively. The relative positions of these subunits are depicted in both complexes. Note that in TFIID, the TBP-binding lobe was poorly resolved19,20 and therefore the second copy of only the Taf6 HEAT repeats and Taf5 NTD is shown.
Extended Data Fig. 5 Interactions between histone-fold pairs in SAGA deviate from the nucleosome pattern already at the four-helix bundle preceding the weak association between Spt3 and Taf10.
The binding between the first two histone-fold pairs along the spiral (left), Taf6–Taf9 with Taf12–Ada1, shows little difference from the nucleosome, possibly reflecting the fact that these histone folds have the highest sequence similarity to histones. As in the nucleosome analogue, hydrophobic interactions have a very important role in this binding. By contrast, the four-helix bundle preceding the association between Spt3 and the Spt7–Taf10 pair, linking Taf12–Ada1 to Spt7–Taf10, is maintained almost solely by hydrogen and electrostatic bonds. Only side chains of residues that participate in the interactions are shown.
Extended Data Fig. 7 TFIIA-dependent TBP release from SAGA.
a, Experimental set-up for pull-down assays that investigate TBP and DNA interactions with SBP-tagged SAGA. b, Gel-shift assay as presented in Fig. 4b but using a longer (100-bp) Cy5-labelled DNA: TATA-containing promoter (TATAL), a TATA-like DNA probe (TATA-likeL) and a non-specific DNA probe (TATA-lessL). The asterisk indicates non-specific DNA association with SAGA. c, Cy5-labelled DNA was almost exclusively double-stranded. Annealed Cy5-labelled DNA used in gel-shift and pull-down assays was loaded on a polyacrylamide 12% gel. Single stranded Cy5-labelled TATA DNA was loaded for comparison. d, Summary of mass-spectrometry data for the major shifted DNA band (Fig. 4, lane 3) showing that it contains both TBP and TFIIA as its major constituents. The spectral count divided by the length in amino acids (PSM/AA) of each protein was normalized with respect to PSM/AA of TBP (raw data are provided in Supplementary Table 1). e, Summary of mass-spectrometry data of purified SAGA. To estimate stoichiometry, PSM/AA was calculated for each protein and normalized with respect to PSM/AA of subunit Tra1 (raw data are provided in Supplementary Table 2). Experiments were repeated three to four times.
Extended Data Fig. 8 Flexibility and anchoring of the enzymatic modules.
a, Two-dimensional classes of SAGA side views revealing different positions of the enzymatic HAT and DUB modules. Bottom, approximate positions of the modules are highlighted in yellow for the HAT module and in blue for the DUB module. The HAT module seems more diffuse and flexible than the DUB module, whose position fluctuates less. b, Docking of the DUB crystal structure (PDB: 6AQR) into the 15 Å resolution cryo-EM map reconstructed from a small class of particles that probably share a similar position for the DUB. c, Domain organization of the Sgf73 subunit. d, The Sgf73 linker tethers DUB to the central module. The linker (S73L) connecting the N-terminal end (ND), which is part of the DUB, and the anchoring domain (S73A) embedded in the central module, includes a SCA7 domain but additionally includes roughly 90 amino acids predicted to be unstructured. All residues that cross-link36 to the Sgf73 linker can be mapped on the surface of SAGA. These residues are depicted with light-blue spheres. A putative path for the 164-residue-long linker is delineated (dashed) according to the cross-linking sites and scarce traces of the linker found in the cryo-EM map. The top insert shows the density that forms contact between DUB and the central module. This contact point is located next to the Spt20 SEP and Taf5 LisH domains and is the only trace of the flexible Sgf73 linker in our maps. e, Two helical domains (ochre) anchor the HAT module at the surface of the Taf6 HEAT repeats (red). f, These helical domains are assigned to Ada3. Domain-deletion analysis identified two domains in Ada3 and one in Ada2 that serve to dock HAT on SAGA and whose deletion results in the release of intact HAT from SAGA36. Fit between the maps and secondary structure predictions (PSIPRED 3.3) or 3D structure of the Ada3 and Ada2 domains is presented in addition to available cross-linking data between these domains and the Taf6 HEAT repeats.
Extended Data Fig. 9 Protein loops and tails contribute to an intricate network of interactions and to bridges between the central module and Tra1.
a, Bridge established by the loop connecting the second and third helices of the cSpt3 histone fold (brown). b, The loop between helices α2 and α3 of the cSpt3 histone fold inserts between two helices of Tra1. Aromatic stacking interactions are formed between Spt3 F294 of this loop and F2404 and F2422 of Tra1. The map density for these three phenylalanine residues is depicted. c, The C-terminal tail of Taf9 (R101–L139) and the short preceding loop illustrate the importance of regions lacking secondary structure elements in SAGA. These parts of Taf9 interact with many subunits and domains, including the Spt3 C′ tail, Spt20 bridge domain, Taf12 histone fold, Ada1, Sgf73 anchoring domain, Taf6 histone fold, Spt7 histone fold, Taf5 WD40, Taf5 NTD and Taf10 histone fold.
Supplementary information
Supplementary Figure 1
Original scanned gels used to produce Figure 4b and Extended Data Figure 7b-c.
Supplementary Data 1
Mass spectrometry analysis of the shifted band in gel shift assays.
Supplementary Data 2
Mass spectrometry analysis of purified SAGA.
Video 1: A tour into SAGA structure
The video shows the EM maps and the derived SAGA structure. It first depicts the central position of the Taf5 WD40 propeller that coordinates the Taf6 HEAT repeats, the Taf5-NTD and the histone-fold octamer. The TBP binding site at the periphery of the deformed octamer is then shown in detail. The video ends by highlighting the connections between the Tra1 and Main lobes as well as the incorporation into the central domain of the Sgf73 subunit which connects to the DUB enzymatic module.
Video 2: The deformed histone-fold octamer in SAGA compared to the symmetrical histone octamer of the nucleosome
The video shows the helix-by-helix alignment of the SAGA histone-fold octamer to the histone octamer and morphs between the aligned and original conformations to highlight the changes.
Rights and permissions
About this article
Cite this article
Papai, G., Frechard, A., Kolesnikova, O. et al. Structure of SAGA and mechanism of TBP deposition on gene promoters. Nature 577, 711–716 (2020). https://doi.org/10.1038/s41586-020-1944-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-1944-2
This article is cited by
-
The NuA4 histone acetyltransferase: variations on a theme of SAGA
Nature Structural & Molecular Biology (2023)
-
Regulation of the RNA polymerase II pre-initiation complex by its associated coactivators
Nature Reviews Genetics (2023)
-
The structure of the NuA4–Tip60 complex reveals the mechanism and importance of long-range chromatin modification
Nature Structural & Molecular Biology (2023)
-
Functional implications of paralog genes in polyglutamine spinocerebellar ataxias
Human Genetics (2023)
-
Conformational landscape of the yeast SAGA complex as revealed by cryo-EM
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.