Abstract
Acute myeloid leukaemia (AML) is a heterogeneous disease characterized by transcriptional dysregulation that results in a block in differentiation and increased malignant self-renewal. Various epigenetic therapies aimed at reversing these hallmarks of AML have progressed into clinical trials, but most show only modest efficacy owing to an inability to effectively eradicate leukaemia stem cells (LSCs)1. Here, to specifically identify novel dependencies in LSCs, we screened a bespoke library of small hairpin RNAs that target chromatin regulators in a unique ex vivo mouse model of LSCs. We identify the MYST acetyltransferase HBO1 (also known as KAT7 or MYST2) and several known members of the HBO1 protein complex as critical regulators of LSC maintenance. Using CRISPR domain screening and quantitative mass spectrometry, we identified the histone acetyltransferase domain of HBO1 as being essential in the acetylation of histone H3 at K14. H3 acetylated at K14 (H3K14ac) facilitates the processivity of RNA polymerase II to maintain the high expression of key genes (including Hoxa9 and Hoxa10) that help to sustain the functional properties of LSCs. To leverage this dependency therapeutically, we developed a highly potent small-molecule inhibitor of HBO1 and demonstrate its mode of activity as a competitive analogue of acetyl-CoA. Inhibition of HBO1 phenocopied our genetic data and showed efficacy in a broad range of human cell lines and primary AML cells from patients. These biological, structural and chemical insights into a therapeutic target in AML will enable the clinical translation of these findings.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The shRNA screen sequencing data have been deposited to the NCBI Sequence Archieve under the accession number GSE120813. Crystal structure data for HBO1–BPRF2 in complex with WM-3835 and acetyl-CoA have been submitted to the PDB under accession numbers 6MAJ (WM-3835) and 6MAK (acetyl-CoA). Source Data are provided for Figs. 1–4. Any other relevant data are available from the corresponding author upon reasonable request.
Code availability
All code used in this study is publically available.
References
Dawson, M. A. The cancer epigenome: concepts, challenges, and therapeutic opportunities. Science 355, 1147–1152 (2017).
Thomas, D. & Majeti, R. Biology and relevance of human acute myeloid leukemia stem cells. Blood 129, 1577–1585 (2017).
Wang, T. et al. Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic Ras. Cell 168, 890–903 (2017).
Tzelepis, K. et al. A CRISPR dropout screen identifies genetic vulnerabilities and therapeutic targets in acute myeloid leukemia. Cell Reports 17, 1193–1205 (2016).
Zuber, J. et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478, 524–528 (2011).
Ying, Q. L. et al. The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 (2008).
Fong, C. Y. et al. BET inhibitor resistance emerges from leukaemia stem cells. Nature 525, 538–542 (2015).
Grimwade, D., Ivey, A. & Huntly, B. J. Molecular landscape of acute myeloid leukemia in younger adults and its clinical relevance. Blood 127, 29–41 (2016).
Tsherniak, A. et al. Defining a cancer dependency map. Cell 170, 564–576 (2017).
Kueh, A. J., Dixon, M. P., Voss, A. K. & Thomas, T. HBO1 is required for H3K14 acetylation and normal transcriptional activity during embryonic development. Mol. Cell. Biol. 31, 845–860 (2011).
Doyon, Y. et al. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. Cell 21, 51–64 (2006).
Sauer, T. et al. MYST2 acetyltransferase expression and histone H4 Lysine acetylation are suppressed in AML. Exp. Hematol. 43, 794–802 (2015).
Weinert, B.T. et al. Time-resolved analysis reveals rapid dynamics and broad scope of the CBP/p300 acetylome. Cell 174, 231–244 (2018).
Saksouk, N. et al. HBO1 HAT complexes target chromatin throughout gene coding regions via multiple PHD finger interactions with histone H3 tail. Mol. Cell 33, 257–265 (2009).
Shi, J. et al. Discovery of cancer drug targets by CRISPR–Cas9 screening of protein domains. Nat. Biotechnol. 33, 661–667 (2015).
Krivtsov, A. V. et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature 442, 818–822 (2006).
Mupo, A. et al. A powerful molecular synergy between mutant nucleophosmin and Flt3-ITD drives acute myeloid leukemia in mice. Leukemia 27, 1917–1920 (2013).
Alharbi, R. A., Pettengell, R., Pandha, H. S. & Morgan, R. The role of HOX genes in normal hematopoiesis and acute leukemia. Leukemia 27, 1000–1008 (2013).
Ballabio, E. & Milne, T. A. Epigenetic control of gene expression in leukemogenesis: cooperation between wild type MLL and MLL fusion proteins. Mol. Cell. Oncol. 1, e955330 (2014).
Skucha, A. et al. MLL-fusion-driven leukemia requires SETD2 to safeguard genomic integrity. Nat. Commun. 9, 1983 (2018).
Church, M., Smith, K. C., Alhussain, M. M., Pennings, S. & Fleming, A. B. Sas3 and Ada2(Gcn5)-dependent histone H3 acetylation is required for transcription elongation at the de-repressed FLO1 gene. Nucleic Acids Res. 45, 4413–4430 (2017).
Shen, W. et al. Solution structure of human Brg1 bromodomain and its specific binding to acetylated histone tails. Biochemistry 46, 2100–2110 (2007).
Huber, F. M. et al. Histone-binding of DPF2 mediates its repressive role in myeloid differentiation. Proc. Natl Acad. Sci. USA 114, 6016–6021 (2017).
Dann, G. P. et al. ISWI chromatin remodellers sense nucleosome modifications to determine substrate preference. Nature 548, 607–611 (2017).
Baell, J. B. et al. Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth. Nature 560, 253–257 (2018).
Majewski, I. J. et al. Polycomb repressive complex 2 (PRC2) restricts hematopoietic stem cell activity. PLoS Biol. 6, e93 (2008).
Voss, A. K., Collin, C., Dixon, M. P. & Thomas, T. Moz and retinoic acid coordinately regulate H3K9 acetylation, Hox gene expression, and segment identity. Dev. Cell 17, 674–686 (2009).
Kinkel, S. A. et al. Jarid2 regulates hematopoietic stem cell function by acting with polycomb repressive complex 2. Blood 125, 1890–1900 (2015).
Keniry, A. et al. Setdb1-mediated H3K9 methylation is enriched on the inactive X and plays a role in its epigenetic silencing. Epigenetics Chromatin 9, 16 (2016).
Dai, Z. et al. edgeR: a versatile tool for the analysis of shRNA-seq and CRISPR-Cas9 genetic screens. F1000Res. 3, 95 (2014).
Shechter, D., Dormann, H. L., Allis, C. D. & Hake, S. B. Extraction, purification and analysis of histones. Nat. Protocols 2, 1445–1457 (2007).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Brumatti, G. et al. HoxA9 regulated Bcl-2 expression mediates survival of myeloid progenitors and the severity of HoxA9-dependent leukemia. Oncotarget 4, 1933–1947 (2013).
Tchasovnikarova, I. A. et al. Epigenetic silencing by the HUSH complex mediates position-effect variegation in human cells. Science 348, 1481–1485 (2015).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 3 (2011).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Lavallée, V. P. et al. The transcriptomic landscape and directed chemical interrogation of MLL-rearranged acute myeloid leukemias. Nat. Genet. 47, 1030–1037 (2015).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995).
Wu, D. et al. ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics 26, 2176–2182 (2010).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Orlando, D. A. et al. Quantitative ChIP-seq normalization reveals global modulation of the epigenome. Cell Reports 9, 1163–1170 (2014).
Ramírez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Robinson, J. T. et al. Integrative Genomics Viewer. Nat. Biotechnol. 29, 24–26 (2011).
Tao, Y., Zhong, C., Zhu, J., Xu, S. & Ding, J. Structural and mechanistic insights into regulation of HBO1 histone acetyltransferase activity by BRPF2. Nucleic Acids Res. 45, 5707–5719 (2017).
Waterman, D. G. et al. Diffraction-geometry refinement in the DIALS framework. Acta Crystallogr. D 72, 558–575 (2016).
Kabsch, W. Xds. Acta Crystallogr. D 66, 125–132 (2010).
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. D 69, 1204–1214 (2013).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D 68, 352–367 (2012).
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997).
Gilan, O. et al. Functional interdependence of BRD4 and DOT1L in MLL leukemia. Nat. Struct. Mol. Biol. 23, 673–681 (2016).
The Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368, 2059–2074 (2013).
Bagger, F. O. et al. BloodSpot: a database of gene expression profiles and transcriptional programs for healthy and malignant haematopoiesis. Nucleic Acids Res. 44, D917–D924 (2016).
Acknowledgements
We thank C. Lovitt, J. Wingerd, S. Jackson and E. Allan for their technical contributions to this project. The work in the Dawson, Blewitt and Burns laboratories was supported by the Cancer Council Victoria Venture Grant Scheme, and Dawson laboratory work was supported by project grant funding from the National Health and Medical Research Council of Australia (1085015). We thank the following funders for fellowship and grant support: Leukaemia Foundation Australia senior fellowship, Cancer Council Victoria Dunlop Fellopship and Howard Hughes Medical Institute international research scholarship (M.A.D.); Victoria Cancer Agency early-career (L.M.) and mid-career (E.Y.N.L.) fellowships; CSL Centenary fellowship (S.-J.D.), Snowdome Foundation (P.Y.), Maddie Riewoldt’s Vision Foundation (Y.-C.C.), Bellberry-Viertel Senior Medical Research Fellowship (M.E.B), Novo Nordisk Foundation Hallas Møller Fellowship NNF14OC0008541 (C.C.), National Health and Medical Research Council of Australia through project grants 1081421 (J.B.B. and T.T.), 575558, 1084248 (A.K.V. and T.T.), research fellowship 1081421 (A.K.V.) and postgraduate scholarship (K.-L.C.). Salary support for M.Z., M.d.S., H.F., C.C., P.S.K., P.A.S., I.P.S. and B.J.M. was provided by the Cancer Therapeutics CRC, funded through the Australian Government’s Cooperative Research Centre programme. The Novo Nordisk Foundation Center for Protein Research is supported financially by the Novo Nordisk Foundation (grant agreement NNF14CC0001). This work was made possible through the Victorian State Government Operation Infrastructure Support and Australian National Health and the Medical Research Council Research Institute Infrastructure Support Scheme. We thank the Australian Synchrotron and beamline scientists for help with data collection; this research was undertaken in part using the MX2 beamline at the Australian Synchrotron and made use of the ACRF detector.
Author information
Authors and Affiliations
Contributions
M.A.D. developed the overall concept behind the study and supervised the project with important advice from L.M., M.E.B., C.J.B., A.K.V., T.T., C.C., F.G. and B.J.M. The manuscript was written by L.M., B.J.M. and M.A.D., with input from all the authors. L.M. performed the majority of the experiments with help from J.A., E.Y.N.L., Y.-C.C., C.-F.W., M.M.Y., P.Y., K.-L.C., K.K., M.S.B., M.L.B., J.L., Y.Y., R.A.B., T.T., H.F., M.d.S., N.N., P.S.K., M.Z., V.M.A. and C.C.-P. Critical reagents and interpretation of the research findings were provided by A.K.V., F.G., S.-J.D., P.A.S., C.C.-P., T.S.P., J.B.B., T.T., O. Dovey, G.S.V., C.C. and I.P.S. The shRNA library was designed and built by L.J.G., T.W., J.L., J.C. and M.E.B. A.H. and C.C. performed mass spectrometry analysis of histone acetylation sites. Protein production, SPR and structural studies were done by S.N., R.S., M.L.D., O. Dolezal, J.N., B.R. and T.S.P. WM-3835 was developed by J.B.B., D.J.L. and N.N. with analytical support by Y.S.
Corresponding author
Ethics declarations
Competing interests
M.A.D. has been a member of advisory boards for CTX CRC, Storm Therapeutics, Celgene and Cambridge Epigenetix. The Dawson, Thomas and Voss laboratories receive research funding from CTx CRC. The remaining authors declare no competing financial interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Peer review information Nature thanks Stefan Knapp and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
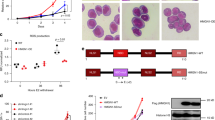
Extended Data Fig. 1 HBO1 is an AML-specific dependency.
a, RNA interference screen multidimensional scaling plot of shRNA sequences from bulk leukaemic blasts (bulk) and LSCs over 14 days of screening. Screens were performed in duplicate (R1 and R2). b, Expression of Ezh2, Lsd1 or Prmt5 mRNA, relative to non-targeting shRNA in LSCs that express shRNAs. n = 3.Mean + s.e.m. c, Negative-selection competition assays in bulk leukaemic blasts and LSCs that express shRNAs against Ezh2, Lsd1, Prmt5 or Dot1l. n = 3. Mean + s.e.m. Validation of the shRNAs against Dot1l have previously been published54. d. Expression of Hbo1 mRNA, relative to non-targeting shRNA, in LSCs that express shRNAs. n = 3. Mean + s.e.m.
Extended Data Fig. 2 Expression levels of HBO1.
a. Box plot of HBO1 expression levels in cancer types from The Cancer Genome Atlas (TCGA)55. The upper limit, centre and lower limit of each box denotes the upper quartile, median and lower quartile of the data, respectively. Cohort abbreviations refer to TCGA study abbreviations (https://gdc.cancer.gov/resources-tcga-users/tcga-code-tables/tcga-study-abbreviations). b, Dot plot of levels of HBO1 expression in normal haematopoietic and AML cells from BloodSpot56; horizontal line indicates mean expression.
Extended Data Fig. 3 HBO1 depletion increases apoptosis, cell-cycle arrest and myeloid differentiation in mouse LSCs.
a, Negative-selection competition assays in LSCs that express shRNAs against Moz, Qkf or Hbo1. n = 3. Mean + s.e.m. b, Negative-selection competition assays in LSCs that express Cas9, transduced with sgRNAs targeting Moz or Qkf. n = 3. Mean + s.e.m. c, Expression of members of the HBO1 complex, relative to non-targeting shRNA in LSCs that express shRNAs targeting individual members of the complex. n = 3. Mean + s.e.m. d, Apoptosis of LSCs that express shRNAs targeting Hbo1. n = 3. Mean + s.e.m. e, Cell-cycle profile of LSCs that express shRNAs targeting Hbo1. n = 3. Mean + s.e.m. f, Surface expression of GR1 and CD11b in LSCs that express shRNAs against Hbo1. n = 3. g. Apoptosis of LSCs that express Cas9 and sgRNAs targeting Hbo1. n = 3. Mean + s.e.m. h, Surface expression of KIT (also known as CD117) in LSCs that express sgRNAs targeting Hbo1. Representative of n = 2 biological replicates.
Extended Data Fig. 4 In vivo depletion of HBO1 increases disease latency.
a, Kaplan–Meier curves of NSG mice transplanted with quinary MLL–AF9 leukaemic cells that express shRNAs targeting Hbo1 or Moz. n = 6 mice per group. b, Schematic of wild-type and Hbo1-mutant alleles with numbered black boxes representing exons. Genotyping primers are indicated (adapted from ref. 10). c, Kaplan–Meier curves of C57BL/6 mice injected with 106 Hbo1flox/+ Mx1-cre primary MLL–AF9 leukaemic cells. Mice were treated with saline or pIpC. n = 12 mice per group. Right, representative genotyping of recipient bone marrow at end point. d, Kaplan–Meier curves of C57BL/6 mice injected with Moz+/+, Moz+/− or Moz−/− MLL–AF9 leukaemic cells. n = 5 mice per group. e, Kaplan–Meier curves of BALB/c mice injected with Moz+/+ or Moz−/− HOXA9 and MEIS1 leukaemic cells. n = 5 mice per group.
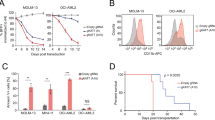
Extended Data Fig. 5 HBO1 is a dependency in various subtypes of AML.
a, Negative-selection competition assays in human AML cell lines that express Cas9 and sgRNAs targeting HBO1. n = 3. Mean + s.e.m. Driver mutations are in parentheses. b, Immunoblot of HBO1 and H3K14ac in OCI-AML3 cells that express Cas9 and sgRNAs targeting HBO1. n = 3. Mean + s.e.m. c, Cell-cycle profile of OCI-AML3 cells that express Cas9 and sgRNAs targeting HBO1. n = 3. Mean + s.e.m. d, Apoptosis of OCI-AML3 cells that express Cas9 and sgRNAs targeting HBO1. n = 3. Mean + s.e.m. e, Surface expression of CD11b in OCI-AML3 cells that express Cas9 and sgRNAs targeting HBO1. n = 3.
Extended Data Fig. 6 HBO1 dependency in other cancers.
Negative-selection competition assays in human cancer cell lines that express Cas9 and sgRNAs targeting HBO1. n = 3. Mean + s.e.m. Cancer type is in parentheses.
Extended Data Fig. 7 HBO1 depletion increases myeloid signature and decreases global H3K14ac.
a, Barcode plot evaluating changes in myeloid development signature after HBO1 depletion with Hbo1 e11.1 and Hbo1 e12.2 sgRNAs in LSCs. n = 3. b, Bar plot of changes in gene expression after Hbo1 deletion in LSCs, ranked by expression levels. Green bars show the top 25 most-downregulated genes after Hbo1 deletion. c, HOXA9 and HOXA10 mRNA expression in Molm13 and OCI-AML3 cells that express Cas9 and sgRNA targeting HBO1. n = 3. Mean ± s.e.m. d, Surface expression of CD11b in LSCs that overexpress Hoxa9 or Hoxa10, and sgRNAs targeting Hbo1. e, Immunoblot of overexpressed HOXA9 or HOXA10 in LSCs that express Cas9. Representative of n = 3 biological replicates. f, Hoxa9- and Hoxa10-rescue negative-selection competition assays. Representative of n = 3 biological replicates. g, ChIP–seq profiles of H3K14ac and RNA Pol-II at the Pbx3 locus in LSCs that express Cas9 and an sgRNA targeting Hbo1. Representative of n = 3 biological replicates. h, RNA Pol-II travelling ratio distribution for highly expressed genes, divided according to H3K14ac levels from ChIP–seq. i, Surface expression of GR1 in LSCs that overexpress sgRNAs targeting Smarca5. Representative of n = 3 biological replicates.
Extended Data Fig. 8 WM-3835 inhibits cell growth and HOXA expression in AML.
a, KAT biochemical and SPR values for WM-3835 compared to WM-1119. Biochemical assay was done at 1 μM acetyl-CoA, the Km of HBO1. b, Proliferation assays of human AML cells treated with 1 μM WM-2474. n = 3. Mean + s.e.m. c, Cellular H3K14ac biomarker assay dose–response curves for WM-3835 (blue) and WM-1119 (red). n = 6. Mean ± s.e.m. d, Growth inhibition assays of the MLL–AF9 AML cell line Molm13 treated with WM-3835 at the doses indicated. Boxes represent minimum and maximum values. n = 11. Mean ± s.e.m. e, Cell-cycle profile of OCI-AML3 cells treated with WM-3835 or vehicle. n = 3. Mean + s.e.m. f, Apoptosis of OCI-AML3 cells treated with WM-3835 or vehicle. n = 3. Mean + s.e.m. g, Surface expression of CD11b in OCI-AML3 cells treated with WM-3835 or vehicle. n = 3. h, HOXA10 mRNA expression in LSCs and Molm13 cells treated with WM-3835 or vehicle. n = 3. Mean + s.e.m. i, HOXA9 and HOXA10 mRNA expression in OCI-AML3 cells treated with WM-3835. n = 3. Mean + s.e.m. j, Hoxa9 and Hoxa10 rescue proliferation assays with 1 μM WM-3835 in LSCs. n = 3. Mean ± s.e.m.
Extended Data Fig. 9 HBO1 crystal structure.
a, Data collection and refinement statistics of WM-3835 HBO1–BRPF2 co-crystal structure. b, WM-3835 binding site in HBO1–BRPF2. WM-3835 is shown in silver with element colouring and the omit electron density map, contoured to 3σ, is shown in green. c, Overlay of WM-3835 and acetyl-CoA (purple with element colouring), showing that WM-3835 binds in the acetyl-CoA binding site of HBO1. d, Ribbon diagram of HBO1–BRPF2 showing WM-3835 bound to the acetyl-CoA binding site. e, Space-filling model showing WM-3835 (yellow with element colouring) in the acetyl-CoA binding pocket of HBO1–BRPF2.
Extended Data Fig. 10 High in vitro metabolism and poor in vivo oral exposure of WM-3835.
a, Proliferation assays of human AML cell lines treated with 1 μM WM-3835. b, WM-3835 demonstrates high clearance in both human and mouse liver microsome assays. The use of dual cofactors (UDPGA and NADPH) results in an increased rate of clearance in human liver microsomes, which is consistent with glucuronidation having a role in the clearance of this compound. c, BALB/c female mice were dosed with WM-3835 at 100 mg kg−1 taken by mouth twice a day, formulated in 20% PEG400/10% solutol or vehicle. Four hours after the third dose, blood samples were collected. An average total drug concentration of 1,860 nM was observed. The free drug level was determined to be 2.6 nM after accounting for mouse plasma protein binding (fraction unbound 0.0014). This free drug level was considered too low to affect the acetylation of H3 at K14, on the basis of the in vitro H3K14ac cellular biomarker data (Fig. 4). An additional chromatographic peak eluting earlier than WM-3835 was detected in the plasma samples from the treatment group. Subsequent analyses using predicted multiple-reaction monitoring and accurate mass measurement indicated that this peak is likely to be a glucuronide conjugate of WM-3835, consistent with the in vitro metabolism data.
Supplementary information
Supplementary Information
This file contains supplementary methods detailing the chemistry for WM-3835.
Supplementary Figure
Contains scans of uncropped images of western blot gels.
Supplementary Table 1
Number of shRNAs depleted by more than 5-fold in leukaemic stem cell and blast cell populations from screen.
Supplementary Table 2
A list of acetylation sites quantified in core histones. Site-specific acetylation of core histones was quantified in HBO1 knockout and control cells using SILAC-based mass spectrometry. The data shows median SILAC ratio (HBO1 knockout/Control) from three independent biological replicates.
Supplementary Table 3
shRNA, sgRNA and mRNA primer sequences.
Rights and permissions
About this article
Cite this article
MacPherson, L., Anokye, J., Yeung, M.M. et al. HBO1 is required for the maintenance of leukaemia stem cells. Nature 577, 266–270 (2020). https://doi.org/10.1038/s41586-019-1835-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1835-6
This article is cited by
-
Aging-induced MCPH1 translocation activates necroptosis and impairs hematopoietic stem cell function
Nature Aging (2024)
-
Guiding the HBO1 complex function through the JADE subunit
Nature Structural & Molecular Biology (2024)
-
RAB27B-regulated exosomes mediate LSC maintenance via resistance to senescence and crosstalk with the microenvironment
Leukemia (2024)
-
Sam68 is a druggable vulnerability point in cancer stem cells
Cancer and Metastasis Reviews (2024)
-
Chromosomal copy number amplification-driven Linc01711 contributes to gastric cancer progression through histone modification-mediated reprogramming of cholesterol metabolism
Gastric Cancer (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.