Abstract
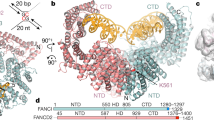
The Fanconi anaemia (FA) pathway repairs DNA damage caused by endogenous and chemotherapy-induced DNA crosslinks, and responds to replication stress1,2. Genetic inactivation of this pathway by mutation of genes encoding FA complementation group (FANC) proteins impairs development, prevents blood production and promotes cancer1,3. The key molecular step in the FA pathway is the monoubiquitination of a pseudosymmetric heterodimer of FANCD2–FANCI4,5 by the FA core complex—a megadalton multiprotein E3 ubiquitin ligase6,7. Monoubiquitinated FANCD2 then recruits additional protein factors to remove the DNA crosslink or to stabilize the stalled replication fork. A molecular structure of the FA core complex would explain how it acts to maintain genome stability. Here we reconstituted an active, recombinant FA core complex, and used cryo-electron microscopy and mass spectrometry to determine its structure. The FA core complex comprises two central dimers of the FANCB and FA-associated protein of 100 kDa (FAAP100) subunits, flanked by two copies of the RING finger subunit, FANCL. These two heterotrimers act as a scaffold to assemble the remaining five subunits, resulting in an extended asymmetric structure. Destabilization of the scaffold would disrupt the entire complex, resulting in a non-functional FA pathway. Thus, the structure provides a mechanistic basis for the low numbers of patients with mutations in FANCB, FANCL and FAAP100. Despite a lack of sequence homology, FANCB and FAAP100 adopt similar structures. The two FANCL subunits are in different conformations at opposite ends of the complex, suggesting that each FANCL has a distinct role. This structural and functional asymmetry of dimeric RING finger domains may be a general feature of E3 ligases. The cryo-electron microscopy structure of the FA core complex provides a foundation for a detailed understanding of its E3 ubiquitin ligase activity and DNA interstrand crosslink repair.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Cryo-EM maps generated during this study have been deposited in the Electron Microscopy Data Bank with accession codes EMD-10290 (FA core complex consensus), EMD-10291 (focused classification top region), EMD-10292 (focused classification middle region), EMD-10293 (focused classification base region) and EMD-10294 (subcomplex). Models generated during this study have been deposited in the Protein Data Bank (PDB) with accession codes 6SRI (FA core complex) and 6SRS (subcomplex). Native mass spectrometry data are available from figshare at https://doi.org/10.6084/m9.figshare.9692192. Crosslinking mass spectrometry data have been deposited in the PRIDE database with accession code PXD014282. All other data are available from the authors upon reasonable request.
References
Crossan, G. P. & Patel, K. J. The Fanconi anaemia pathway orchestrates incisions at sites of crosslinked DNA. J. Pathol. 226, 326–337 (2012).
Schlacher, K., Wu, H. & Jasin, M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51–BRCA1/2. Cancer Cell 22, 106–116 (2012).
Nalepa, G. & Clapp, D. W. Fanconi anaemia and cancer: an intricate relationship. Nat. Rev. Cancer 18, 168–185 (2018).
Knipscheer, P. et al. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science 326, 1698–1701 (2009).
Smogorzewska, A. et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell 129, 289–301 (2007).
Rajendra, E. et al. The genetic and biochemical basis of FANCD2 monoubiquitination. Mol. Cell 54, 858–869 (2014).
Garcia-Higuera, I. et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell 7, 249–262 (2001).
Huang, Y. et al. Modularized functions of the Fanconi anemia core complex. Cell Rep. 7, 1849–1857 (2014).
Swuec, P. et al. The FA core complex contains a homo-dimeric catalytic module for the symmetric mono-ubiquitination of FANCI–FANCD2. Cell Rep. 18, 611–623 (2017).
Pace, P. et al. FANCE: the link between Fanconi anaemia complex assembly and activity. EMBO J. 21, 3414–3423 (2002).
van Twest, S. et al. Mechanism of ubiquitination and deubiquitination in the Fanconi anemia pathway. Mol. Cell 65, 247–259 (2017).
Walden, H. & Deans, A. J. The Fanconi anemia DNA repair pathway: structural and functional insights into a complex disorder. Annu. Rev. Biophys. 43, 257–278 (2014).
Nakane, T., Kimanius, D., Lindahl, E. & Scheres, S. H. Characterisation of molecular motions in cryo-EM single-particle data by multi-body refinement in RELION. eLife 7, e36861 (2018).
Bai, X. C., Rajendra, E., Yang, G., Shi, Y. & Scheres, S. H. Sampling the conformational space of the catalytic subunit of human γ-secretase. eLife 4, e11182 (2015).
Cole, A. R., Lewis, L. P. C. & Walden, H. The structure of the catalytic subunit FANCL of the Fanconi anemia core complex. Nat. Struct. Mol. Biol. 17, 294–298 (2010).
Kowal, P., Gurtan, A. M., Stuckert, P., D’Andrea, A. D. & Ellenberger, T. Structural determinants of human FANCF protein that function in the assembly of a DNA damage signaling complex. J. Biol. Chem. 282, 2047–2055 (2007).
Huang, A. et al. Symmetry and asymmetry of the RING–RING dimer of Rad18. J. Mol. Biol. 410, 424–435 (2011).
Mattiroli, F. et al. RNF168 ubiquitinates K13–15 on H2A/H2AX to drive DNA damage signaling. Cell 150, 1182–1195 (2012).
Zhang, M. et al. Chaperoned ubiquitylation—crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP–Ubc13–Uev1a complex. Mol. Cell 20, 525–538 (2005).
Brzovic, P. S., Rajagopal, P., Hoyt, D. W., King, M. C. & Klevit, R. E. Structure of a BRCA1–BARD1 heterodimeric RING–RING complex. Nat. Struct. Biol. 8, 833–837 (2001).
Buchwald, G. et al. Structure and E3-ligase activity of the Ring–Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 25, 2465–2474 (2006).
Passmore, L. A. et al. Structural analysis of the anaphase-promoting complex reveals multiple active sites and insights into polyubiquitylation. Mol. Cell 20, 855–866 (2005).
Tang, X. et al. Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell 129, 1165–1176 (2007).
Gordon, S. M., Alon, N. & Buchwald, M. FANCC, FANCE, and FANCD2 form a ternary complex essential to the integrity of the Fanconi anemia DNA damage response pathway. J. Biol. Chem. 280, 36118–36125 (2005).
Polito, D. et al. The carboxyl terminus of FANCE recruits FANCD2 to the Fanconi Anemia (FA) E3 ligase complex to promote the FA DNA repair pathway. J. Biol. Chem. 289, 7003–7010 (2014).
Zimmerman, E. S., Schulman, B. A. & Zheng, N. Structural assembly of cullin-RING ubiquitin ligase complexes. Curr. Opin. Struct. Biol. 20, 714–721 (2010).
Neveling, K., Endt, D., Hoehn, H. & Schindler, D. Genotype–phenotype correlations in Fanconi anemia. Mutat. Res. 668, 73–91 (2009).
Sari, D. et al. The MultiBac baculovirus/insect cell expression vector system for producing complex protein biologics. Adv. Exp. Med. Biol. 896, 199–215 (2016).
Weissmann, F. et al. biGBac enables rapid gene assembly for the expression of large multisubunit protein complexes. Proc. Natl Acad. Sci. USA 113, E2564–E2569 (2016).
Hill, C. H. et al. Activation of the endonuclease that defines mRNA 3′ ends requires incorporation into an 8-subunit core cleavage and polyadenylation factor complex. Mol. Cell 73, 1217–1231.e11 (2019).
Sato, K., Toda, K., Ishiai, M., Takata, M. & Kurumizaka, H. DNA robustly stimulates FANCD2 monoubiquitylation in the complex with FANCI. Nucleic Acids Res. 40, 4553–4561 (2012).
Russo, C. J. & Passmore, L. A. Ultrastable gold substrates for electron cryomicroscopy. Science 346, 1377–1380 (2014).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Fernandez-Leiro, R. & Scheres, S. H. W. A pipeline approach to single-particle processing in RELION. Acta Crystallogr. D 73, 496–502 (2017).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Scheres, S. H. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 415, 406–418 (2012).
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014).
García-Nafría, J., Lee, Y., Bai, X., Carpenter, B. & Tate, C. G. Cryo-EM structure of the adenosine A2A receptor coupled to an engineered heterotrimeric G protein. eLife 7, e35946 (2018).
Stewart, P. L., Burnett, R. M., Cyrklaff, M. & Fuller, S. D. Image reconstruction reveals the complex molecular organization of adenovirus. Cell 67, 145–154 (1991).
He, J., Schmid, M. F., Zhou, Z. H., Rixon, F. & Chiu, W. Finding and using local symmetry in identifying lower domain movements in hexon subunits of the herpes simplex virus type 1 B capsid. J. Mol. Biol. 309, 903–914 (2001).
Rossmann, M. G. & Blow, D. M. Detection of sub-units within crystallographic asymmetric unit. Acta Crystallogr. D 15, 24–31 (1962).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Yang, J. et al. The I-TASSER suite: protein structure and function prediction. Nat. Methods 12, 7–8 (2015).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Holm, L. & Sander, C. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 20, 478–480 (1995).
Mourão, A., Nager, A. R., Nachury, M. V. & Lorentzen, E. Structural basis for membrane targeting of the BBSome by ARL6. Nat. Struct. Mol. Biol. 21, 1035–1041 (2014).
Delorenzi, M. & Speed, T. An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics 18, 617–625 (2002).
Zimmermann, L. et al. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 430, 2237–2243 (2018).
Wood, C. W. & Woolfson, D. N. CCBuilder 2.0: powerful and accessible coiled-coil modeling. Protein Sci. 27, 103–111 (2018).
Nookala, R. K., Hussain, S. & Pellegrini, L. Insights into Fanconi anaemia from the structure of human FANCE. Nucleic Acids Res. 35, 1638–1648 (2007).
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D 67, 355–367 (2011).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Goddard, T. D. et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
van de Waterbeemd, M. et al. High-fidelity mass analysis unveils heterogeneity in intact ribosomal particles. Nat. Methods 14, 283–286 (2017).
Rose, R. J., Damoc, E., Denisov, E., Makarov, A. & Heck, A. J. High-sensitivity Orbitrap mass analysis of intact macromolecular assemblies. Nat. Methods 9, 1084–1086 (2012).
Gault, J. et al. High-resolution mass spectrometry of small molecules bound to membrane proteins. Nat. Methods 13, 333–336 (2016).
Taverner, T. et al. Subunit architecture of intact protein complexes from mass spectrometry and homology modeling. Acc. Chem. Res. 41, 617–627 (2008).
Chen, Z. A. et al. Architecture of the RNA polymerase II–TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 29, 717–726 (2010).
Kolbowski, L., Mendes, M. L. & Rappsilber, J. Optimizing the parameters governing the fragmentation of cross-linked peptides in a tribrid mass spectrometer. Anal. Chem. 89, 5311–5318 (2017).
Lenz, S., Giese, S. H., Fischer, L. & Rappsilber, J. In-search assignment of monoisotopic peaks improves the identification of cross-linked peptides. J. Proteome Res. 17, 3923–3931 (2018).
Giese, S. H., Fischer, L. & Rappsilber, J. A study into the collision-induced dissociation (CID) behavior of cross-linked peptides. Mol. Cell. Proteomics 15, 1094–1104 (2016).
Fischer, L. & Rappsilber, J. Quirks of error estimation in cross-linking/mass spectrometry. Anal. Chem. 89, 3829–3833 (2017).
Naydenova, K. & Russo, C. J. Measuring the effects of particle orientation to improve the efficiency of electron cryomicroscopy. Nat. Commun. 8, 629 (2017).
Buetow, L. & Huang, D. T. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 17, 626–642 (2016).
Knipscheer, P. & Sixma, T. K. Protein–protein interactions regulate Ubl conjugation. Curr. Opin. Struct. Biol. 17, 665–673 (2007).
Metzger, M. B., Pruneda, J. N., Klevit, R. E. & Weissman, A. M. RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta 1843, 47–60 (2014).
Linares, L. K., Hengstermann, A., Ciechanover, A., Müller, S. & Scheffner, M. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc. Natl Acad. Sci. USA 100, 12009–12014 (2003).
Alpi, A. F., Pace, P. E., Babu, M. M. & Patel, K. J. Mechanistic insight into site-restricted monoubiquitination of FANCD2 by Ube2t, FANCL, and FANCI. Mol. Cell 32, 767–777 (2008).
Acknowledgements
We thank T. Nakane, J. Zivanov, C. Lau, A. Carter, P. Emsley, G. Murshudov, D. Malinverni and M. Babu for advice and discussions; K. Naydenova, B. Santhanam, G. Dornan, D. Briant, A. Casañal, A. Kumar, M. Carminati, A. Kelley and members of the Passmore laboratory for assistance; G. Cannone, C. Savva and the LMB EM facility, J. Grimmett and T. Darling (LMB scientific computation) for support and J. Shi for assistance with baculovirus. This work was supported by the Medical Research Council, as part of United Kingdom Research and Innovation, MRC file reference number MC_U105192715 (L.A.P.); Deutsche Forschungsgemeinschaft (DFG, 329673113) (J.R.); the Wellcome Trust through a Senior Research Fellowship to J.R. (103139); and the European Research Council grant number 695511-ENABLE (C.V.R). The Wellcome Centre for Cell Biology is supported by core funding from the Wellcome Trust (203149). We acknowledge Diamond Light Source for access to eBIC (proposals EM18091 and EM17434) funded by the Wellcome Trust, MRC and Biotechnology and Biological Sciences Research Council.
Author information
Authors and Affiliations
Contributions
S.S., E.R. and P.A. designed protein expression and purification schemes, performed ubiquitination assays and performed cryo-EM, 3D reconstruction and modelling; S.S. and D.S.C. performed native mass spectrometry; F.O’R. and G.D. performed crosslinking mass spectrometry; S.M. performed hydrogen–deuterium exchange–mass spectrometry; S.H. and S.H.W.S. developed the local symmetry algorithm; C.H.H., C.J.R. and L.A.P. collected cryo-EM data; L.A.P., C.V.R., J.M.S., J.R., S.H.W.S. and K.J.P. supervised the research; L.A.P. conceived the project; and S.S., P.A. and L.A.P. wrote the paper with contributions from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Peer review information Nature thanks Andrew Deans, Xiaodong Zhang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Fig. 1 Recombinant FA core complex activity.
a, Ubiquitination assay analysed by western blot with HA antibody to detect HA-tagged ubiquitin. The migration positions of monoubiquitinated FANCD2 and FANCI are indicated but FANCI is not substantially modified. b, Ubiquitination assay analysed by Coomassie-stained SDS–PAGE to show specific monoubiquitination of FANCD2 K563 by recombinant FA core complex. Wild type (WT), FANCD2(K563R) and FANCI(K525R) (KR) were analysed. A native FA core complex purified from chicken DT40 cells monoubiquinates FANCD2 but does not efficiently monoubiquitinate FANCI6. Therefore, the purified recombinant complex faithfully recapitulates the properties of the native chicken complex. Notably, a purified human complex also did not efficiently monoubiquitinate FANCI, although it did efficiently monoubiquitinate FANCD211. The asymmetry in the FA core complex (see below) reflects this asymmetry in its activity on FANCD2–FANCI. An additional factor or post-translational modification may be required for activation of FANCI monoubiquitination. The ubiquitination assays were repeated at least two times independently with similar results. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 2 Cryo-EM reconstruction of FA core complex, multi-body refinement and assessment of 3D reconstructions after focused refinement.
a, Representative raw micrograph of FA core complex. b, Overall 3D reconstruction of the FA core complex. c, Angular distribution density plot of particles used in the 3D reconstruction of the FA core complex. Every point is a particle orientation and the colour scale represents the normalized density of views around this point. The colour scale runs from 0 (low, blue) to 0.00026 (high, red). The efficiency of orientation distribution67, EOD, was 0.79. d, Estimated local resolution map for FA core complex. e, FSC plot for gold standard refinement. f, Multibody refinement of the FA core complex using three masks (body 1, body 2 and body 3) shown in pink, purple and cyan, respectively. The motions are shown in Supplementary Videos 1 and 2. g, Local resolution maps for reconstructions of top, middle and base regions of FA core complex. The middle region did not substantially change between multi-body refinement and particle subtraction followed by focused classification. The resolution of the base and top regions improved after particle subtraction and focused classification and refinement. h, FSC plot for gold standard refinements. i, Representative density for β-strand and α-helical regions. FANCB–FAAP100 is in the middle region which is better defined than more peripheral regions, including FANCF.
Extended Data Fig. 3 Subunit assignment and arrangement in FA core complex.
a–c, Complexes lacking specific subunits were purified and analysed by cryo-EM. a, Major 2D class averages identified for subcomplexes, compared with those from FA core complex (A-G-B-L-100-C-E-F). Cartoons are shown to depict the subunits visible in the class averages. The symmetric subcomplex identified in the FA core complex preparation is indicated (sym). The A-G-B-L-100 complex (lacking the substrate-recognition module) has similar symmetric 2D classes. Native mass spectrometry revealed a non-uniform subunit stoichiometry. Thus it is likely that the asymmetric assembly represents the complete FA core complex, while the symmetric structure is a subcomplex that co-purifies with the intact complex. The 2D class average of a complex lacking FANCA (G-B-L-100-C-E-F) appeared similar to the complete FA core complex with no obvious missing density. FANCG is probably the partially disordered arm that extends from the central part of the complex since this was missing when FANCG was not present in the complex. b, Coomassie-stained SDS–PAGE analysis of purified subcomplexes. Asterisks indicate contaminant proteins. FANCA did not co-purify with the A-B-L-100 complex but its migration position is indicated on the gel. The purifications were repeated at least two times independently with similar results. For gel source data, see Supplementary Fig. 1. c, Cartoon of FA core complex with subunits labelled. d, Native mass spectrum of recombinant FA core complex showing masses and subunit composition of assigned peak series. We dissociated the FA core complex into subcomplexes during ionization and these species were detected by mass spectrometry. Computational analyses then revealed the proteins present in each of the peaks. The standard deviation in fitting the identified peaks to the charge series is given as the ± error in the measured mass for a given single measurement. This is the error in the fit and not the error in the mass measurement, which is probably an order of magnitude higher due to, for instance, solvation or adduct effects, or heterogeneous post-translational modifications. Hence, the error gives a rough measure of the accuracy of peak assignment, which is impacted by the broadness, symmetry and signal-to-noise ratio of each peak. e, Molecular masses of FA core complex subunits. The expected and measured masses are given, along with phosphorylation sites detected by mass spectrometry. Native mass spectrometry was repeated three times with similar results.
Extended Data Fig. 4 Crosslinking mass spectrometry.
a, Crosslinking mass spectrometry revealed 834 crosslinks (1% FDR) between residues that are in close proximity. Intermolecular crosslinks are coloured in green; intramolecular crosslinks, red; predicted α-helices, orange; predicted β-strands, purple. Asterisks on FANCB mark missense mutations from the Fanconi Anaemia Mutation Database (http://www2.rockefeller.edu/fanconi/) including L43S in a predicted β-strand, P230S at the N terminus of a predicted helix, and L329P in the middle of a predicted β-strand, all in the β-propeller; and L676P, which is predicted to disrupt an α-helix in the C-terminal dimerization domain. b–d, Validation of crosslinking of FA core complex. Crosslinks were mapped onto a homology model of chicken FANCL (b), human FANCF (c) and human FANCE (d). All intramolecular crosslinks within a domain are consistent with the maximum crosslinker length (30 Å between the two Cα). Crosslinks between the domains in FANCL are not consistent with the domain arrangement in the crystal structure of FANCL because there is flexibility or changes in the orientation between the domains in the FA core complex. There are two different conformations of FANCL in the structure: FANCLbase is fully ordered, whereas only the ELF domain is visible in FANCLtop. Mapping the crosslinks onto FANCL from the base (constrained, with all three domains visible) reveals that the distances for some crosslinks between domains are too large to be consistent with the FANCL conformation in the base. By contrast, for FANCL in the top, where only the ELF domain is ordered, the URD and RING domains are likely to be conformationally flexible. Since the URD and RING domains cannot be modelled for FANCLtop, it is not possible to validate these interdomain crosslinks.
Extended Data Fig. 5 Assessment of model fit in maps and modelling of coiled coils.
a–c, FSC plots of maps versus model for top (a), middle (b) and base (c) regions. d, Low-resolution cryo-EM map of FA core complex (transparent surface) with models placed in the map. Asterisks represent density that was not visible at high resolution and was not interpreted. This may represent FANCA and additional parts of the substrate-recognition module. e, MARCOIL51 prediction for the best heptad phase in the long helices of FANCB and FAAP100. f, Predicted coiled-coil model by CCbuilder 2.053 for the FANCB and FAAP100 long helices. Crosslinks detected for these helices in the FA core complex crosslinking mass spectrometry are indicated with blue lines.
Extended Data Fig. 6 HDX-MS on FANCB and FAAP100.
a–d, Difference plots for FANCB showing peptides that are protected (negative) or exposed (positive) upon binding of additional subunit(s) for B-L-100 vs B-100 (a), G-B-L-100 vs B-L-100 (b), A-G-B-L-100 vs G-B-L-100 (c) and FA core complex vs. G-B-L-100 (d). Exchange of hydrogens in FANCB residues 429–448 was protected after interaction with FANCL, consistent with FANCL being located next to the coiled coil. e–h, Difference plots for FAAP100 showing peptides that are protected (negative) or exposed (positive) upon binding of additional subunit(s) for B-L-100 vs B-100 (e), G-B-L-100 vs B-L-100 (f), A-G-B-L-100 vs. G-B-L-100 (g) and FA core complex vs G-B-L-100 (h). Exchange of hydrogens in FAAP100 residues 448–464 was protected after interaction with FANCL, consistent with FANCL being located next to the coiled coil. For difference plots, triplicate data from four independent colour-coded time points are shown. The significance threshold is indicated by dashed lines. Grey shading indicates the standard deviation of all charge states and replicates per peptide. Sequence coverage is shown in the Supplementary Information.
Extended Data Fig. 7 3D reconstruction of symmetric FA subcomplex and local symmetry refinement.
a, Overall 3D reconstruction of the symmetric FA subcomplex. b, Angular distribution plot of particles used in the 3D reconstruction of the symmetric FA subcomplex. Every point is a particle orientation and the colour scale represents the normalized density of views around this point. The colour scale runs from 0 (low, blue) to 0.00026 (high, red). The efficiency of orientation distribution67, EOD, was 0.65. c, Estimated local resolution map for symmetric FA subcomplex. d, FSC plot for gold standard refinement. e, Local symmetry pipeline for reconstruction of the symmetric FA subcomplex (see Methods). This reconstruction could not be improved with C2 symmetry, probably because of local flexibility. f, FSC plot for gold standard refinement shown for the subcomplex reconstruction before and after local symmetry refinement. The circular panels show representative densities before and after local symmetry refinement.
Extended Data Fig. 8 Model for FA subcomplex.
a, Model of FA subcomplex shown as cartoon representations of subunits fit into the cryo-EM map. b, Model of FA subcomplex shown as a surface representation of the combined models. Two views are shown down the two-fold symmetric axis. c, Comparison of FA subcomplex and complete FA core complex in the same orientations. Both models are shown in cartoon representation. Subunits are coloured as in Fig. 1e. d, Modelling of a fully symmetric FA core complex containing two copies of every subunit. Left, the second copy of FANCG (cartoon) from the FA subcomplex was modelled onto the structure of the FA core complex (surface representation). This second FANCG clashes with the FANCB β-propeller in the base (asterisks). Thus, it is likely that upon binding of the substrate-recognition module, rearrangement of the β-propellers of FANCB and FAAP100 prevents binding of a second copy of FANCG. (Right) A second copy of substrate-recognition module (FANCC–FANCE–FANCF; cartoon) is modelled in the top region of the FA core complex by combining the models of FANCC–FANCE–FANCF and FANCLbase from FA core complex followed by superimposing FANCLbase on FANCLtop. There is a clash (asterisks) between the modelled FANCL (cartoon) and FAAP100 β-propeller (surface representation). These data suggest that a fully symmetric complex does not readily form. In agreement with this, there was no evidence for any classes containing two copies of C-E-F in any of our EM analyses of the FA core complex. e, f, The symmetric FA subcomplex (A-G-B-L-100) does not readily associate with purified substrate-recognition module (C-E-F) to form the asymmetric FA core complex. e, The 2D class averages of A-G-B-L-100 mixed with C-E-F compared with complete FA core complex, FA subcomplex and A-G-B-L-100 subcomplex. A-G-B-L-100 was mixed with C-E-F in molar ratios of 1:1 and 1:2 for 1 h at 4 °C before cryo-plunging. Only the symmetric A-G-B-L-100 subcomplex was observed and there was no additional density for C-E-F. Panels for FA core complex, subcomplex and A-G-B-L-100 are replicated from Extended Data Fig. 3a. f, Pull-down assay of C-E-F using tagged A-G-B-L-100 (Strep II tagged). Left, the Coomassie Blue-stained gel of purified C-E-F (with Strep II tag, after 3C cleavage of tag and after removal of 3C protease). Tagged A-G-B-L-100 was immobilized on StrepTactin resin and incubated with purified C-E-F at a 1:1 molar ratio. After washing, only a small amount of C-E-F remains bound to the beads. Negative controls (A-G-B-L-100 only and C-E-F only) are shown in the middle panel. Asterisk indicates a contaminant protein. The pulldown experiment was repeated two times independently with similar results. Since C-E-F does not efficiently bind A-G-B-L-100, these experiments suggest that these species are unlikely to be in equilibrium in solution. Previous genetic and biochemical data show that FANCC, FANCE and FANCF are important for monoubiquitination of the FANCD2–FANCI substrate. Together, these data provide evidence that the asymmetric complex is the relevant, functional, physiological entity.
Extended Data Fig. 9 Structural comparison of E3 ligases.
a, There is a strong precedence for dimerization of RING/U-box domain E3 ubiquitin ligases68,69,70. RING/U-box E3s exist both as homo- and heterodimeric complexes, for example, Rad18–Rad18, CHIP–CHIP, RNF8–RNF8, BRCA1–BARD1, RING1b–BMI1 and Hdm2–Hdmx17,18,19,20,21,71. Structures of homo- and heterodimeric RING/U-box E3 ligases are shown here with the RING/U-box in orange. Surprisingly, these E3s display functional and structural asymmetry: in all the dimers listed above, only one protomer binds to an E2 enzyme. The homodimeric CHIP E3 ligase has a strikingly asymmetric structure that clearly demonstrates why only one U-box binds E2 enzyme19. The FANCL RING subunit is also an asymmetric dimer within the FA core complex and it is possible that only one of these binds E2. However, unlike the smaller E3 s, the FANCL RING fingers are not near each other. Together, this suggests that asymmetric dimerization may be a general feature of RING E3s. b, Comparison of FA core complex with cullin–RING ubiquitin ligases (CRLs). Many large complexes are predominantly helical suggesting that α-helices are commonly used as building blocks for complexes. In addition, β-propellers often mediate protein–protein interactions. The CRL complexes and FA core complex are long and extended with substrate-recognition (green), scaffold (yellow) and RING (orange) subunits residing in three different regions of the structure. However, the structural details of these complexes differ. Interestingly, the activities of some multisubunit RING-containing E3 ligases including APC/C and CRL complexes are stimulated by dimerization22,23. Thus, dimerization may underpin physiological ubiquitination activity in many E3 ligases. c, d, Ubiquitin discharge assay, in which free lysine is used instead of the FANCD2–FANCI substrate. In these experiments, the FA core complex is incubated with E1, E2, ubiquitin and free lysine. If FANCL is active without a substrate, ubiquitin will be conjugated to lysine, resulting in a shift in its molecular weight; however, if substrate binding is required to activate the E3 ligase activity (for example, through allosteric changes), this will not occur. Coomassie gels of reaction products were run in non-reducing (c) and reducing (100 mM DTT) conditions (d). Ubiquitin is transferred to free lysine as shown by the increase in molecular weight of ubiquitin as well as a decrease in intensity of the E2–ubiquitin band when compared to the lane containing no free lysine. Thus, substrate binding is not required for activity. Reducing conditions do not eliminate the UBE2T–ubiquitin conjugate, as previously shown72. Additionally, DNA is not required for FA core complex E3 ligase activity on free lysines, suggesting that DNA activates the substrate, not the E3. The ubiquitin discharge assays were repeated three times independently with similar results (c, d). e, Distributions of patient mutations are indicated on the FA core complex by heat map colouring of subunits and in percentage. f, Ubiquitination assay using several subcomplexes (Extended Data Fig. 3b) and the full FA core complex, analysed by western blot with HA antibody to detect HA-tagged ubiquitin. The migration positions of monoubiquitinated FANCD2 and FANCI are indicated but FANCI is not substantially modified, as in Extended Data Fig. 1a. All complexes have similar activities but isolated FANCL is less active. This assay was repeated at least two times independently with similar results. For gel source data, see Supplementary Fig. 1.
Supplementary information
Supplementary Information
This file contains Supplementary Figure 1 and Supplementary Data 1 and 2.
Video 1:
Multi-body refinement, motion 1. Principal component analysis of multi-body refinement results revealed two major motions. This video shows motion in the FA core complex for principal component 1, swiveling of the base.
Video 2:
Multi-body refinement, motion 2. Principal component analysis of multi-body refinement results revealed two major motions. This video shows motion in the FA core complex for principal component 2, bending along the long axis.
Video 3:
3D reconstruction of complete FA core complex and three sub-domains (top, middle and base). The consensus 3D reconstruction is shown (grey surface), followed by the focused refinement map for the top (yellow), multi-body refined map for the middle (cyan) and the focused refinement map for the base (magenta) regions.
Video 4:
Models for FA core complex subunits fit into the cryoEM map of the complete complex. Models are shown in cartoon representation, coloured by subunit as indicated, in a composite cryoEM map of the top, middle and base regions (transparent).
Video 5:
Models for FA subunits fit into the cryoEM map of the subcomplex. Models are shown in cartoon representation, coloured by subunit as indicated in Supplementary Video 4 and Fig. 1e, in the cryoEM map (transparent).
Video 6:
Conformational differences in the two copies of FANCB and FAAP100. Morph of FANCB-FAAP100 between the symmetric FA subcomplex and asymmetric complete FA core complex models. Models are shown in cartoon representation (coloured as in Supplementary Video 4 and Fig. 1e).
Rights and permissions
About this article
Cite this article
Shakeel, S., Rajendra, E., Alcón, P. et al. Structure of the Fanconi anaemia monoubiquitin ligase complex. Nature 575, 234–237 (2019). https://doi.org/10.1038/s41586-019-1703-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1703-4
This article is cited by
-
FAAP100 is required for the resolution of transcription-replication conflicts in primordial germ cells
BMC Biology (2023)
-
The DNA-damage kinase ATR activates the FANCD2-FANCI clamp by priming it for ubiquitination
Nature Structural & Molecular Biology (2022)
-
The structure-specific endonuclease complex SLX4–XPF regulates Tus–Ter-induced homologous recombination
Nature Structural & Molecular Biology (2022)
-
Retention time prediction using neural networks increases identifications in crosslinking mass spectrometry
Nature Communications (2021)
-
Structure of the FA core ubiquitin ligase closing the ID clamp on DNA
Nature Structural & Molecular Biology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.