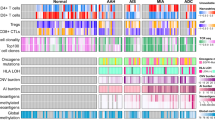
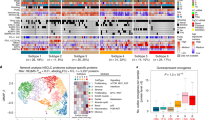
Abstract
Early detection and treatment are critical for improving the outcome of patients with cancer1. Understanding the largely uncharted biology of carcinogenesis requires deciphering molecular processes in premalignant lesions, and revealing the determinants of the intralesional immune reaction during cancer development. The adaptive immune response within tumours has previously been shown to be strongest at the earliest stage of carcinoma2,3. Here we show that immune activation and immune escape occur before tumour invasion, and reveal the relevant immune biomarkers of the pre-invasive stages of carcinogenesis in the lung. We used gene-expression profiling and multispectral imaging to analyse a dataset of 9 morphological stages of the development of lung squamous cell carcinoma, which includes 122 well-annotated biopsies from 77 patients. We identified evolutionary trajectories of cancer and immune pathways that comprise (1) a linear increase in proliferation and DNA repair from normal to cancerous tissue; (2) a transitory increase of metabolism and early immune sensing, through the activation of resident immune cells, in low-grade pre-invasive lesions; (3) the activation of immune responses and immune escape through immune checkpoints and suppressive interleukins from high-grade pre-invasive lesions; and, ultimately, (4) the activation of the epithelial–mesenchymal transition in the invasive stage of cancer. We propose that carcinogenesis in the lung involves a dynamic co-evolution of pre-invasive bronchial cells and the immune response. These findings highlight the need to develop immune biomarkers for early detection as well as immunotherapy-based chemopreventive approaches for individuals who are at high risk of developing lung cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Gene expression data are available in the Gene Expression Omnibus database with accession number GSE33479.
Code availability
Code is available on Github at https://github.com/Precancer/SCC.
References
Goldberg, S. W., Mulshine, J. L., Hagstrom, D. & Pyenson, B. S. An actuarial approach to comparing early stage and late stage lung cancer mortality and survival. Popul. Health Manag. 13, 33–46 (2010).
Bindea, G. et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 39, 782–795 (2013).
Mlecnik, B. et al. Histopathologic-based prognostic factors of colorectal cancers are associated with the state of the local immune reaction. J. Clin. Oncol. 29, 610–618 (2011).
Herbst, R. S., Morgensztern, D. & Boshoff, C. The biology and management of non-small cell lung cancer. Nature 553, 446–454 (2018).
De Koning, H., Van Der Aalst, C., Ten Haaf, K. & Oudkerk, M. PL02.05 effects of volume CT lung cancer screening: mortality results of the NELSON randomised-controlled population based trial. J. Thorac. Oncol. 13, S185 (2018).
Umar, A., Dunn, B. K. & Greenwald, P. Future directions in cancer prevention. Nat. Rev. Cancer 12, 835–848 (2012).
Kensler, T. W. et al. Transforming cancer prevention through precision medicine and immune-oncology. Cancer Prev. Res. 9, 2–10 (2016).
Slaughter, D. P., Southwick, H. W. & Smejkal, W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer 6, 963–968 (1953).
Kerr, K. M. Pulmonary preinvasive neoplasia. J. Clin. Pathol. 54, 257–271 (2001).
Galon, J. et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 313, 1960–1964 (2006).
Galon, J., Angell, H. K., Bedognetti, D. & Marincola, F. M. The continuum of cancer immunosurveillance: prognostic, predictive, and mechanistic signatures. Immunity 39, 11–26 (2013).
Pagès, F. et al. International validation of the consensus immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet 391, 2128–2139 (2018).
Pagès, F. et al. In situ cytotoxic and memory T cells predict outcome in patients with early-stage colorectal cancer. J. Clin. Oncol. 27, 5944–5951 (2009).
Fridman, W. H., Pagès, F., Sautès-Fridman, C. & Galon, J. The immune contexture in human tumours: impact on clinical outcome. Nat. Rev. Cancer 12, 298–306 (2012).
Gentles, A. J. et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 21, 938–945 (2015).
Teixeira, V. H. et al. Deciphering the genomic, epigenomic, and transcriptomic landscapes of pre-invasive lung cancer lesions. Nat. Med. 25, 517–525 (2019).
Wu, S., Powers, S., Zhu, W. & Hannun, Y. A. Substantial contribution of extrinsic risk factors to cancer development. Nature 529, 43–47 (2016).
Angelova, M. et al. Evolution of metastases in space and time under immune selection. Cell 175, 751–765 (2018).
Mlecnik, B. et al. The tumor microenvironment and immunoscore are critical determinants of dissemination to distant metastasis. Sci. Transl. Med. 8, 327ra26 (2016).
Galon, J. & Bruni, D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov. 18, 197–218 (2019).
Gandhi, L. et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N. Engl. J. Med. 378, 2078–2092 (2018).
Reck, M. et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N. Engl. J. Med. 375, 1823–1833 (2016).
Brahmer, J. et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N. Engl. J. Med. 373, 123–135 (2015).
Herbst, R. S. et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 387, 1540–1550 (2016).
Antonia, S. J. et al. Overall survival with durvalumab after chemoradiotherapy in stage III NSCLC. N. Engl. J. Med. 379, 2342–2350 (2018).
Eggermont, A. M. M. et al. Adjuvant pembrolizumab versus placebo in resected stage III melanoma. N. Engl. J. Med. 378, 1789–1801 (2018).
Forde, P. M. et al. Neoadjuvant PD-1 blockade in resectable lung cancer. N. Engl. J. Med. 378, 1976–1986 (2018).
Mascaux, C. et al. Evolution of microRNA expression during human bronchial squamous carcinogenesis. Eur. Respir. J. 33, 352–359 (2009).
Bota, S. et al. Follow-up of bronchial precancerous lesions and carcinoma in situ using fluorescence endoscopy. Am. J. Respir. Crit. Care Med. 164, 1688–1693 (2001).
Breuer, R. H. et al. The natural course of preneoplastic lesions in bronchial epithelium. Clin. Cancer Res. 11, 537–543 (2005).
Acknowledgements
We thank P. Pierard, J. Faber, M. Bruyneel, S. Bensliman and O. Bauwens, who performed bronchoscopy sampling; A.-P. Meert and T. Berghmans, who recruited the patients from Jules Bordet Institute; C. Decoster, B. Martin and J.-F. Godart, who contributed to biopsy collection; Y. Cleuter, A. Vandenbroecke, J.-F. Laes and L. Lafontaine, who shared their technical expertise; B. Grisart, P. Martiat and C. Sotiriou, who opened access to their laboratory; and all the patients who generously participated in this study. This research was supported by the Fund of Medical Scientific Research (Belgium), the National Fund of Scientific Research-Télévie campaign and the ASBL ‘Les Amis de l’Institut Jules Bordet’. C.M. was a research fellow from the National Fund of Scientific Research (Belgium) and was supported by a grant from the Louisiana Chapter of the National Lung Cancer Partnership (USA). J.B. was supported by a LUNGevity Foundation Career Development Award. This work was also supported by grants from the National Cancer Institute of France (INCa), the Canceropole Ile de France, INSERM, La Ligue Contre le Cancer, the Cancer research for personalized medicine (CARPEM), Paris Alliance of Cancer Research Institutes (PACRI) and LabEx Immuno-oncology.
Reviewer information
Nature thanks Lynette Marie Sholl and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
C.M., A.B., J.-P.S. and J.G. designed the study. C.M., G.A., A.H. and V.N. collected, handled and classified the samples, and collected the clinical data. C.M., G.A., B.B. and F.R. performed the experiments. C.M., A.V., G.A. and B.B. acquired the data. M.A., C.M., A.V., J.B., K.H., K.W.-G., A.B., J.-P.S., A.S. and J.G. analysed and interpreted the data. M.A., C.M., J.B. and K.H. performed the statistical analyses. C.M., J.B., J.-P.S. and J.G. acquired the funding required for the study. C.M., A.B., J.-P.S, A.S. and J.G. supervised the project. C.M., M.A. and J.G. drafted the manuscript. All authors performed a critical revision of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare competing financial interests: J.G. is co-founder of HalioDx biotech company. A.S. is an employee of Johnson and Johnson. C.M. received honoraria for expertise and speaker roles, being on the advisory board or attending meetings, from Boehringher Ingelheim, Lilly, Bristol-Myers Squibb, AstraZeneca, Roche, Kephren and Novartis. European Patent Application EP19305434.3 was filed on 2 April 2019 by Inserm, Université Paris Descartes, Université Paris Diderot, Sorbonne Université, Université Aix-Marseille, APHM and Université Libre de Bruxelles, and European Patent Application EP19305535.7 was filed on 26 April 2019 by Inserm, Université Paris Descartes, Université Paris Diderot, Sorbonne Université, Université Aix-Marseille, APHM and Université Libre de Bruxelles. The inventors designation is in progress.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Methodology for studying tumorigenesis.
a, Across nine morphological stages of the development of lung SCC, freshly frozen samples were assayed for gene-expression profiling. The two methodological axes are visualized separately; one flow chart focuses on the detection of gene-expression patterns (left) and one flow chart focuses on the in-depth immune characterization (right) from gene expression and multispectral imaging. On the basis of gene co-expression, molecular phenotypes and gene modules were defined and functionally characterized. Immune-gene signatures and deconvolution methods were used for quantitative assessment of different immune-cell types. Relevant immune cells were investigated in more depth using multiplex immunohistochemistry and multispectral imaging. b, Bronchoscopy of in situ carcinoma tissue with white light (left) and with 400-nm wavelength illumination, under which the bronchial epithelium appears in green (right). Although it is difficult to distinguish the in situ carcinoma from normal tissue under white light, the in situ carcinoma displays a lack of green fluorescence compared to the normal epithelial tissue under fluorescence bronchoscopy.
Extended Data Fig. 2 Four molecular steps in carcinogenesis of lung SCC.
The heat map shows genes that are associated with developmental stages identified using a linear mixed-effects model. Annotation bars are included for cancer history, sex and smoking history, all of which were used as fixed factors for the linear model, along with patient information (which was used as a random effect). Gene expression discerned four molecular groups: normal, low grade, high grade and SCC, on the basis of semi-supervised hierarchical clustering. Normal tissue with normal fluorescence, hypofluorescent normal tissue and hyperplasia lesions were subsumed under the category of normal tissue (stages 0, 1 and 2); metaplasia, mild dysplasia and moderate dysplasia were grouped as low grade (stages 3, 4 and 5); severe dysplasia and carcinoma in situ comprised the high-grade category; and the invasive stage was singled out as SCC.
Extended Data Fig. 3 Hallmarks of cancer.
a, Single-sample gene-set enrichment analysis was performed on the full expression profile using cancer-hallmark gene definitions from mSigDb (v.6.2), independently of the gene modules. The heat map visualizes the enrichment scores from the single-sample gene-set enrichment analysis, in which the samples were ordered by their average enrichment scores for each molecular group individually. Only the cancer hallmarks that were significant with respect to the over-representation analysis of the gene modules in Fig. 1b are shown for validation. Three hallmark definitions associated with the ascending module at the highest adjusted P values (P > 0.003) are shown here (these are not shown in Fig. 1b): PI3K_AKT_MTOR_SIGNALING (proliferation), UV_RESPONSE_UP (DNA damage, confirmed by the UV_RESPONSE_DN in the descending module) and HYPOXIA (pathway). b, Left, randomized principal component (PC) analysis on the full expression profile shows a gradual continuum of expression changes from stage 0 to stage 8. Middle, randomized principal component analysis on enrichment scores for cancer hallmarks revealed distinct molecular steps. The cancer hallmarks explained up to 76.2% of the sample variability with the first two principal components (middle) as opposed to 24.9% variability explained by the full expression profile (left). Right, based on the principal component rotations, the hallmarks of proliferation, immune system, metabolism and the epithelial–mesenchymal transition each contribute to defining the developmental stages, as observed by their different directions of variability. c, Increase in expression of key genes and chemokines involved in the epithelial–mesenchymal transition, together with genes that overlap with the hallmark signature of the epithelial–mesenchymal transition and the ‘SCC increase’ gene module. The differential expression analysis of chemokines related to the epithelial–mesenchymal transition considered the confounding factors of smoking status, cancer history, sex, age and inter-patient variability. No differential expression across the developmental stages was found for CXCL1 and CXCL6, whereas the expression of CXCR2 was significantly increased in both the high-grade and SCC lesions. d, Only CXCR4 had a significant increase specific to SCC that was not observed in low- and high-grade lesions.
Extended Data Fig. 4 Comparison of deconvolution approaches.
a, Immune estimates derived from the CIBERSORT method, using the LM22 gene signature (all 22 cell types are presented) (top) or using on our in-house-developed immune signature (HD signature) (bottom). b, Symmetric correlation matrix of the average immune-cell abundance per developmental stage (Spearman correlation), estimated with CIBERSORT using the LM22 gene signature (left) and the HD signature (right). c, Immune co-regulation and immune-status shift derived from CIBERSORT estimates using the HD signature. d, Comparison of the mcpCounter, TIMER, EPIC and xCell methods for expression-based interrogation of the tumour immune infiltrates.
Extended Data Fig. 5 Intrapatient immune heterogeneity.
a, Stacked bars illustrate the relative cellular abundance of different immune-cell types estimated with CIBERSORT, in patients sampled for multiple grades. There are similar profiles for samples from the same grade, independent of the patient. b, Single-sample gene-set enrichment analysis was performed on the HD immune-cell signatures. The heat map represents one-dimensional clustering by immune-cell type, in which the samples of each molecular group were ordered by their average enrichment score. c, A chord diagram links the samples derived from the same patient. The order of the samples is preserved from b.
Extended Data Fig. 6 Quantitative and spatial immune characterization through immunohistochemistry and multispectral imaging.
a, Immunohistochemistry (IHC) quantification of the immune checkpoints CTLA4, IDO1, TIGIT and TIM3. Each of the tested markers was validated in SCC tissue (top). P values are derived from a non-parametric one-tailed Mann–Whitney U test used to validate increase in SCC compared to normal tissue. b, Comparison of PD-L1 densities between the stroma of normal tissue (stage 0) and SCC (stage 8), derived from multiplex immunohistochemistry. c, Clustering of normalized immunohistochemistry expression. d, A methodology for spatial analysis of multispectral imaging data. A whole slide is reconstructed from the individual images. On the basis of the tissue categorization, the images are masked to exclude the blank areas. Immune-cell densities are calculated as the number of cells per tissue area (m2). Spatial localization is analysed within the selected region of interest. e, Representative examples of CK−PD-L1+ in both SCC and severe dysplasia. Single-positive PD-L1 cells (CK−PD-L1+) were generally immune cells that were located in the stroma, with morphological similarities to infiltrating macrophages. f, We calculated the area between the theoretical and the empirical curve because deviations between the two can indicate clustering or segregation patterns (see Fig. 4b, bottom) to confirm that epithelial cells segregate from CD3 T cells in high-grade lesions, independently from the distance threshold of 25 μm.
Extended Data Fig. 7 Graphical overview of lung carcinogenesis.
Schematic illustrating the main stages of carcinogenesis for lung SCC.
Extended Data Fig. 8 Quality control of microarray data.
a, Distribution of the relative abundance of each probe; that is, the ratio between the red and green colour intensity (Cy5/Cy3) for all probes across all patients (log2-transformed). b, Gene-expression distribution for each hybridization and amplification run (left). Using randomized principal component analysis, the samples were projected on the first two principal components and highlighted with different colours on the basis of their hybridization run (top right) and amplification run (bottom right). c, Classification of the samples is based on the expression of sex-chromosome genes, along with colour annotation for the sex of the patient.
Extended Data Fig. 9 Detection of gene co-expression modules.
a, Expression of 4,734 genes that were associated with the nine developmental stages (linear mixed-C. effects model) is illustrated for each of the seven expression trajectories (that is, gene modules), detected using weighted network analysis of gene co-expression. b, Only 18 out of 4,734 genes were not assigned to a gene module and did not follow any of the seven illustrated expression patterns. c, A weighted network of genes is constructed by raising the adjacency matrix to a power. The value of the power for soft thresholding was chosen to be 12, as the lowest power term at which the network approximately fits a scale-free topology (red line R2 ≤ 0.85). The horizontal red line shows the squared correlation (R2) cut-off of 0.85 recommended by the scale-free topology criterion. d, We randomly split the full dataset into a reference and test set, and evaluated the module preservation across the respective networks (n = 50 samples). The Zsummary statistic (top) provides evidence that the observed value of the preservation statistic is significantly higher than expected by chance (strong evidence if Zsummary > 10; weak-to-moderate if Zsummary > 2 and < 10; no evidence if Zsummary < 2). The grey module is unclassified, and expectedly showed no preservation (Zsummary < 2). All of the modules were preserved between the reference and the test datasets (Zsummary > 10), except in the SCC increase module (Zsummary = 9.1) which is also the smallest module with gene expression in small number of samples after resampling (an increase only in SCC). The median rank-preservation statistic showed, independently of the module size, that there is stronger preservation for all modules compared to the grey unclassified set of genes (bottom). e, The dendrogram derived from hierarchal clustering of the topology overlap matrix dissimilarity measure of the full dataset is shown in the top panel. The modules are defined on the basis of this dendrogram using a dynamic tree cut (top panel). We applied the same parameters for weighted gene-correlation network analysis on a resampled subset of the full dataset (a randomly selected two-thirds of the full dataset). The resampling was performed without replacement, which ensured proportional representation of each developmental stage. Colour rows indicate the module assignments obtained on the full dataset (first row) and on the resampled subsets of samples (n = 50 samples). All of the seven gene modules identified in the full dataset appear in almost every resampling, which indicates that the modules are stable.
Extended Data Fig. 10 Tumour heterogeneity.
a, Single-sample gene-set enrichment analysis using pannormal tissue, and normal lung tissue from HD gene signature. b, Estimates of epithelial-cell abundance derived from the xCell method (see Supplementary Information). c, Expression of the proliferation gene-marker MKI67. d, Expression of the proliferation gene-marker in CK+ cells.
Supplementary information
Supplementary Information
This file contains supplementary text and supplementary table legends for tables S1-S4.
Supplementary Table
Supplementary Table 1 | Clinicopathological characteristics of the cohort summarized per developmental stage.
Supplementary Table
Supplementary Table 2 | A list of all studied samples of the corresponding patients of the cohort. Samples whose blocks could not be analysed with multispectral imaging were marked with X, concerning both the phenotype and the functional panel.
Supplementary Table
Supplementary Table 3 | Functional analysis of differential gene expression. Immune-related Gene Ontology biological processes significantly enriched among differentially regulated genes in low-grade, high-grade, and SCC compared to normal.
Supplementary Table
Supplementary Table 4 | Compendium of immune genes referred to as the HD gene signature.
Rights and permissions
About this article
Cite this article
Mascaux, C., Angelova, M., Vasaturo, A. et al. Immune evasion before tumour invasion in early lung squamous carcinogenesis. Nature 571, 570–575 (2019). https://doi.org/10.1038/s41586-019-1330-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1330-0
This article is cited by
-
Smoking-associated gene expression alterations in nasal epithelium reveal immune impairment linked to lung cancer risk
Genome Medicine (2024)
-
Immunosurveillance encounters cancer metabolism
EMBO Reports (2024)
-
Diagnostic Value of GDF10 for the Tumorigenesis and Immune Infiltration in Lung Squamous Cell Carcinoma
Current Medical Science (2024)
-
Comparative immunological landscape between pre- and early-stage LUAD manifested as ground-glass nodules revealed by scRNA and scTCR integrated analysis
Cell Communication and Signaling (2023)
-
Convergence of YAP/TAZ, TEAD and TP63 activity is associated with bronchial premalignant severity and progression
Journal of Experimental & Clinical Cancer Research (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.