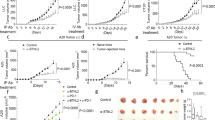
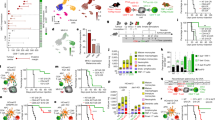
Abstract
Solid tumours are infiltrated by effector T cells with the potential to control or reject them, as well as by regulatory T (Treg) cells that restrict the function of effector T cells and thereby promote tumour growth1. The anti-tumour activity of effector T cells can be therapeutically unleashed, and is now being exploited for the treatment of some forms of human cancer. However, weak tumour-associated inflammatory responses and the immune-suppressive function of Treg cells remain major hurdles to broader effectiveness of tumour immunotherapy2. Here we show that, after disruption of the CARMA1–BCL10–MALT1 (CBM) signalosome complex, most tumour-infiltrating Treg cells produce IFNγ, resulting in stunted tumour growth. Notably, genetic deletion of both or even just one allele of CARMA1 (also known as Card11) in only a fraction of Treg cells—which avoided systemic autoimmunity—was sufficient to produce this anti-tumour effect, showing that it is not the mere loss of suppressive function but the gain of effector activity by Treg cells that initiates tumour control. The production of IFNγ by Treg cells was accompanied by activation of macrophages and upregulation of class I molecules of the major histocompatibility complex on tumour cells. However, tumour cells also upregulated the expression of PD-L1, which indicates activation of adaptive immune resistance3. Consequently, blockade of PD-1 together with CARMA1 deletion caused rejection of tumours that otherwise do not respond to anti-PD-1 monotherapy. This effect was reproduced by pharmacological inhibition of the CBM protein MALT1. Our results demonstrate that partial disruption of the CBM complex and induction of IFNγ secretion in the preferentially self-reactive Treg cell pool does not cause systemic autoimmunity but is sufficient to prime the tumour environment for successful immune checkpoint therapy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All datasets generated during the current study are available from the corresponding authors upon reasonable request. RNA sequencing data have been deposited at the Gene Expression Omnibus (GEO) under accession number GSE129480.
References
Savage, P. A., Leventhal, D. S. & Malchow, S. Shaping the repertoire of tumor-infiltrating effector and regulatory T cells. Immunol. Rev. 259, 245–258 (2014).
Mellman, I., Coukos, G. & Dranoff, G. Cancer immunotherapy comes of age. Nature 480, 480–489 (2011).
Spranger, S. et al. Up-regulation of PD-L1, IDO, and Tregs in the melanoma tumor microenvironment is driven by CD8+ T cells. Sci. Transl. Med. 5, 200ra116 (2013).
Bauer, C. A. et al. Dynamic Treg interactions with intratumoral APCs promote local CTL dysfunction. J. Clin. Invest. 124, 2425–2440 (2014).
Meininger, I. & Krappmann, D. Lymphocyte signaling and activation by the CARMA1-BCL10-MALT1 signalosome. Biol. Chem. 397, 1315–1333 (2016).
Medoff, B. D. et al. Differential requirement for CARMA1 in agonist-selected T-cell development. Eur. J. Immunol. 39, 78–84 (2009).
Molinero, L. L. et al. CARMA1 controls an early checkpoint in the thymic development of FoxP3+ regulatory T cells. J. Immunol. 182, 6736–6743 (2009).
Barnes, M. J. et al. Commitment to the regulatory T cell lineage requires CARMA1 in the thymus but not in the periphery. PLoS Biol. 7, e1000051 (2009).
Brüstle, A. et al. MALT1 is an intrinsic regulator of regulatory T cells. Cell Death Differ. 24, 1214–1223 (2017).
Schmidt-Supprian, M. et al. Differential dependence of CD4+CD25+ regulatory and natural killer-like T cells on signals leading to NF-κB activation. Proc. Natl Acad. Sci. USA 101, 4566–4571 (2004).
Smigiel, K. S. et al. CCR7 provides localized access to IL-2 and defines homeostatically distinct regulatory T cell subsets. J. Exp. Med. 211, 121–136 (2014).
Long, M., Park, S.-G., Strickland, I., Hayden, M. S. & Ghosh, S. Nuclear factor-κB modulates regulatory T cell development by directly regulating expression of Foxp3 transcription factor. Immunity 31, 921–931 (2009).
Oh, H. et al. An NF-κB transcription-factor-dependent lineage-specific transcriptional program promotes regulatory T cell identity and function. Immunity 47, 450–465.e5 (2017).
Vasanthakumar, A. et al. The TNF receptor superfamily-NF-κB axis is critical to maintain effector regulatory T cells in lymphoid and non-lymphoid tissues. Cell Rep. 20, 2906–2920 (2017).
Messina, N. et al. The NF-κB transcription factor RelA is required for the tolerogenic function of Foxp3+ regulatory T cells. J. Autoimmun. 70, 52–62 (2016).
Grinberg-Bleyer, Y. et al. NF-κB c-Rel is crucial for the regulatory T cell immune checkpoint in cancer. Cell 170, 1096–1108.e13 (2017).
Yu, J. et al. Regulation of T-cell activation and migration by the kinase TBK1 during neuroinflammation. Nat. Commun. 6, 6074 (2015).
Jenkins, M. H. et al. Multiple murine BRafV600E melanoma cell lines with sensitivity to PLX4032. Pigment Cell Melanoma Res. 27, 495–501 (2014).
Pierson, W. et al. Antiapoptotic Mcl-1 is critical for the survival and niche-filling capacity of Foxp3+ regulatory T cells. Nat. Immunol. 14, 959–965 (2013).
Overacre-Delgoffe, A. E. et al. Interferon-γ drives Treg fragility to promote anti-tumor immunity. Cell 169, 1130–1141.e11 (2017).
Gewies, A. et al. Uncoupling Malt1 threshold function from paracaspase activity results in destructive autoimmune inflammation. Cell Reports 9, 1292–1305 (2014).
Jaworski, M. et al. Malt1 protease inactivation efficiently dampens immune responses but causes spontaneous autoimmunity. EMBO J. 33, 2765–2781 (2014).
Bornancin, F. et al. Deficiency of MALT1 paracaspase activity results in unbalanced regulatory and effector T and B cell responses leading to multiorgan inflammation. J. Immunol. 194, 3723–3734 (2015).
Nagel, D. et al. Pharmacologic inhibition of MALT1 protease by phenothiazines as a therapeutic approach for the treatment of aggressive ABC-DLBCL. Cancer Cell 22, 825–837 (2012).
Schlauderer, F. et al. Structural analysis of phenothiazine derivatives as allosteric inhibitors of the MALT1 paracaspase. Angew. Chem. Int. Edn Engl. 52, 10384–10387 (2013).
Fontan, L. et al. MALT1 small molecule inhibitors specifically suppress ABC-DLBCL in vitro and in vivo. Cancer Cell 22, 812–824 (2012).
Wang, Y. et al. MALT1 promotes melanoma progression through JNK/c-Jun signaling. Oncogenesis 6, e365 (2017).
Thome, M., Charton, J. E., Pelzer, C. & Hailfinger, S. Antigen receptor signaling to NF-κB via CARMA1, BCL10, and MALT1. Cold Spring Harb. Perspect. Biol. 2, a003004–a003004 (2010).
Le, D. T. et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 372, 2509–2520 (2015).
Rizvi, N. A. et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128 (2015).
Zaretsky, J. M. et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N. Engl. J. Med. 375, 819–829 (2016).
Rubtsov, Y. P. et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity 28, 546–558 (2008).
Rubtsov, Y. P. et al. Stability of the regulatory T cell lineage in vivo. Science 329, 1667–1671 (2010).
Srinivas, S. et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 4 (2001).
Sasaki, Y. et al. Canonical NF-κB activity, dispensable for B cell development, replaces BAFF-receptor signals and promotes B cell proliferation upon activation. Immunity 24, 729–739 (2006).
Dalton, D. K. et al. Multiple defects of immune cell function in mice with disrupted interferon-γ genes. Science 259, 1739–1742 (1993).
Godfrey, V. L., Wilkinson, J. E., Rinchik, E. M. & Russell, L. B. Fatal lymphoreticular disease in the scurfy (sf) mouse requires T cells that mature in a sf thymic environment: potential model for thymic education. Proc. Natl Acad. Sci. USA 88, 5528–5532 (1991).
Egawa, T. et al. Requirement for CARMA1 in antigen receptor-induced NF-κB activation and lymphocyte proliferation. Curr. Biol. 13, 1252–1258 (2003).
Marangoni, F. et al. The transcription factor NFAT exhibits signal memory during serial T cell interactions with antigen-presenting cells. Immunity 38, 237–249 (2013).
Spiess, P. J., Yang, J. C. & Rosenberg, S. A. In vivo antitumor activity of tumor-infiltrating lymphocytes expanded in recombinant interleukin-2. J. Natl. Cancer Inst. 79, 1067–1075 (1987).
Marangoni, F. et al. Tumor tolerance-promoting function of regulatory T cells is optimized by CD28, but strictly dependent on calcineurin. J. Immunol. 200, 3647–3661 (2018).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Acknowledgements
We thank the MGH Pathology Flow Cytometry Core and N. Ali-Akbar for technical assistance. This study was funded by an EMBO fellowship (ALTF534-2015) and a Marie Curie Global Fellowship (750973) (M.D.P.), DFG Fellowships (PR 1652/1-1 to J.N.P and US 116/2-1 to S.M.U), NIH T32 CA207021 (V.M.), a Sara Elizabeth O’Brien Fellowship (F.M.), and Melanoma Research Alliance Senior Investigator Award MRA-348693, NIH AI123349, and the Bob and Laura Reynolds MGH Research Scholar Award (T.R.M.).
Reviewer information
Nature thanks Shimon Sakaguchi and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
M.D.P. initiated, designed, performed and analysed the experiments, and wrote the manuscript. E.Y.K. initiated the project, designed and performed experiments, V.Z. performed histological analyses, S.M.U. performed autoantibody assays, V.M. and F.M. performed Treg cell analyses in lung and skin. F.M. performed in vitro Treg suppression assay. E.C. generated tumour cell lines. M.N.N. and A.-C.V. performed RNA sequencing analyses, B.D.M. provided genetic mouse models, D.S. designed and performed RT–qPCR assay, B.L.C., S.M., J.N.P., R.D.W. and M.L. performed tumour growth studies and survival studies, T.R.M. conceived the study, supervised the project, designed experiments, and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
M.D.P. and T.R.M. have filed a patent application (PCT/US2018/067856) related to the use of MALT1 inhibitors. T.R.M. is a co-founder of Monopteros Therapeutics. All other authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Lymphoproliferative disease after Treg-cell-specific deletion of CARMA1.
a, CARMA1 protein in Treg cells and CD4+ Tconv cells from LNs of Fcre × C1+/+, C1f/+ and C1f/f mice. b, Weight curves (n = 5 per group). c, d, Appearance of 21-day-old mice (c), and their spleens and LNs (d). e, Histological appearance of liver, skin and lung at 21 days of age of indicated mice. Scale bars, 150 μm and 50 μm (insets). f, Kidney, liver and stomach tissue sections of healthy C57BL/6 Rag1−/− mice were reacted with serum from 21-day-old mice of the indicated genotypes, and self-tissue-reactive IgG revealed by anti-mouse IgG staining (green). Nuclei were stained with DAPI (red). In b, *P < 0.05 versus C1+/+ and C1f/+ (two-way ANOVA with Bonferroni post hoc test).
Extended Data Fig. 2 Myeloid cell expansion and effector cytokine secretion by Tconv and Treg cells after Treg-cell-specific deletion of CARMA1.
a–c, Size of the CD11b+ splenic myeloid compartment and proportions of Ly6G+ neutrophils, CD11c+MHC IIhigh dendritic cells (DCs), Ly6Chigh monocytes, Lyc6GlowSSChigh eosinophils, and Ly6ClowSSClow macrophages in Fcre × C1+/+, C1f/+ or C1f/f mice. d, Expression of MHC-I, MHC-II and PD-L1 on splenic myeloid subsets. e, Frequency of CD4+ and CD8+ Tconv cells with a CD44highCD62L− effector memory phenotype in LNs of indicated mice at age 12 and 21 days. f, Effector cytokine expression of Tconv cells from 21-day-old mice after 8-h ex vivo stimulation on anti-CD3/CD28-coated plates. g, LN cellularity. h, Absolute numbers of Treg cells in LNs. i, Co-expression of indicated transcription factors by Treg cells from LNs of indicated mice. j, Expression of CD44 and CD62L by Fcre × C1f/f Treg cells expressing T-bet (green dots), GATA-3 (blue dots), or RORγt (red dots), compared to total C1f/f Treg cells (contour plots). *P < 0.05, **P < 0.01, ***P < 0.001, ***P < 0.0001 (one-way ANOVA with Tukey post hoc test).
Extended Data Fig. 3 Role of CARMA1 in eTreg cell differentiation.
a, Female heterozygous Fcre/+ × C1f/f mice express YFP–Cre and delete C1f/f in half of the Treg cells owing to X-chomosomal location of the Foxp3YFP-cre allele and random X chromosome inactivation, whereas the other half of the Treg cells remains functional. b, Frequency of CD4+ and CD8+ Tconv cells with a CD44highCD62L– effector memory phenotype in peripheral blood of ageing Fcre/+ × C1+/+, C1f/+ or C1f/f mice (n = 4 per group). c, Appearance of spleens and LNs of indicated mice at one year of age. d–f, Frequency and absolute numbers (e) of eTreg cells and expression of FOXP3, indicated markers of eTreg cell differentiation, as well as proliferation marker Ki67, pro-apoptotic protein BIM and anti-apoptotic protein BCL2 by YFP+ cTreg and eTreg cells (d, f) from nine-week-old Fcre/+ × C1+/+, C1f/+ or C1f/f mice. Note that some data on eTreg cells in e and f are also shown in Fig. 1g, h and shown here to facilitate the comparison to cTreg and YFP– Treg cells in g and h. g, h, Frequency and absolute numbers of eTreg cells (g) and eTreg cell markers on YFP– cTreg and eTreg cells (h) from the same mice as shown in d and f. *P < 0.05, **P < 0.01, ***P < 0.001, ***P < 0.0001 (one-way ANOVA with Tukey post hoc test).
Extended Data Fig. 4 In vitro and in vivo suppression, apoptotic rate and exTreg cell formation of CARMA1-deficient Treg cells.
a, CD4+CD45RBhighYFP– Tconv cells and CD4+CD45RBlowYFPbright Treg cells were double-sorted to more than 98% purity from LNs and spleens of Fcre × C1+/+ × ROSA26-stopf/f-YFP mice, which allow for clear differentiation of Cre-expressing Treg cells based on high expression of soluble enhanced yellow fluorescent protein (eYFP) in addition to the YFP–Cre fusion protein. b, YFPbright Treg from Fcre × C1+/+ or Fcre/+ × C1f/+ or C1 f/f mice and CellTrace Violet-labelled Tconv cells from Fcre × C1+/+ mice were co-cultured at indicated ratios for three days in the presence of anti-CD3 antibodies and T-cell-depleted splenocytes, and suppression measured as reduction of Tconv cell proliferation. c, Treg cells of various genotypes and Tconv cells were co-adoptively transferred into Rag-deficient hosts and their respective frequency in peripheral blood was determined eight weeks later. d, CD4+YFP+ Treg cells of indicated genotypes were cultured without exogenous IL-2 on anti-CD3/CD28-coated or uncoated plates for 6 or 18 h and examined for reactivity with annexin V and the viability dye ZombieRed. e, CD4+YFPbright cells were sorted from LNs of one-year-old and Fcre/+ × C1+/+ (or C1 f/+, C1 f/f) × ROSA26-stopf/f-YFP mice and subsequently stained for expression of FOXP3 protein to determine the frequency of FOXP3– exTreg cells. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (two-way ANOVA with Sidak post hoc test in b, c; two-tailed Student’s t-test in d).
Extended Data Fig. 5 Bulk RNA sequencing analysis of YFP+ cTreg and eTreg cells from LNs of Fcre/+ × C1+/+, C1f/+ and C1f/f mice.
a, Scaled expression in cTreg cells (top) and eTreg cells (bottom) of genes differentially expressed (fold change > 2 and Padj < 0.05) between C1+/+ and C1f/f mice. b, High-resolution, fully annotated heat map of eTreg signature genes as shown in Fig. 1j.
Extended Data Fig. 6 Expression of apoptotic regulator genes.
a, Comparison of normalized, log-transformed mRNA expression levels of anti- and pro-apoptotic Bcl2 family genes in YFP+ cTreg and eTreg from Fcre/+ × C1+/+ mice, based on RNA sequencing analyses. b, Comparison of expression of the same genes in cTreg (top) and eTreg (bottom) cells of the indicated genotypes. *Padj < 0.05.
Extended Data Fig. 7 Restoring NF-κB activation in CARMA1-deficient Treg cells.
a, Survival of Fcre × C1+/+ (or C1f/f) × ROSA26-stopf/f-IKK2ca mice that express one allele of IKK2ca after expression of Foxp3cre. b–d, Frequency of CD4+ and CD8+ Tconv cells with a CD44highCD62L– effector memory phenotype in LNs (b), frequency of Treg among CD4+ T cells and of eTreg cells among total Treg cells in LNs (c), and effector cytokine expression of Tconv cells after 8-h ex vivo stimulation on anti-CD3/CD28-coated plates (d) in indicated mice. e, f, Expression of indicated phospho-proteins by YFP+ Treg cells from Fcre/+ × C1+/+ or × C1f/f mice at baseline and after 30 min. Anti-CD3/CD28 in vitro stimulation. Grey solid histograms show unstained control cells. Tconv cells were used as internal controls, and showed no differences (not shown). *P < 0.05, ***P < 0.001, ****P < 0.0001 (log-rank (Mantel–Cox) test in a; one-way ANOVA with Tukey post hoc test in b–d, f).
Extended Data Fig. 8 Role of IFNγ secretion by FOXP3int unstable Treg cells selectively in tumour tissue.
a–c, Frequencies of total YFP+ Treg cells and of YFP+ eTreg cells among CD4+ T cells (a), absolute YFP+ Treg and eTreg cell numbers (b), and normalized FOXP3 expression in YFP+ Treg and eTreg (c) in tdLN and tumour tissue of 18-day-old D4M.3A tumours in female heterozygous Fcre/+ × C1+/+, C1 f/+ or C1 f/f mice. d, tdLN and tumour cellularities. e, Growth of MC38 tumours in female heterozygous Fcre/+ × C1+/+ or C1 f/+ mice. f, g, Tumour-bearing mice were treated with brefeldin A for 5 h, injected intravenously with 3 μg of anti-THY1.2 monoclonal antibodies, and 3 min later collected for direct ex vivo analysis of IFNγ expression in extravascular YFP+ Treg cells in the lung (f) and skin (g). Gates for IFNγ+ cells drawn based on fluorescence-minus-one (FMO) controls. h, Normalized FOXP3 expression in IFNγ+ and IFNγ– Treg cells from tumour tissue. n.d., not detectable. i, Tumour growth in indicated mice implanted with D4M.3A melanoma and19 treated with or without neutralizing anti-IFNγ antibody. j, Frequency of adoptively transferred, YFP+ Treg cells of indicated genotypes in tdLNs of Ifng−/− hosts at day 18 of tumour growth. k–m, Frequency (k, l) and effector cytokine expression (m) of adoptively transferred, YFP+ Treg cells in tumours in Ifng−/− hosts. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (one-way ANOVA with Tukey post hoc test in a–d, f, g; two-tailed Student’s t-test in e, h, j, l, m; two-way ANOVA with Bonferroni post hoc test in i).
Extended Data Fig. 9 Tumour response of CARMA1-deficient Treg cells after restoration of NF-κB activation.
a–d, D4M.3A melanoma cells were implanted into Fcre/+ × C1+/+ (or C1 f/f) × ROSA26-stopf/f-IKK2ca mice to record frequencies of YFP+ Treg and eTreg cells among CD4+ T cells (a) and their normalized FOXP3 expression (b) in tdLNs and tumour tissue, effector cytokine expression by tumour-infiltrating Treg cells (c), and tumour growth (d). e, YFP+ Treg cells were sorted from D4M.3A melanoma tissue and tdLNs after five days of treatment of FcreERT2 × C1+/+ and C1f/f with tamoxifen and analysed for CARMA1 expression by RT–qPCR. f, Tumour growth in female FcreERT2 or FcreERT2/+ × C1+/+ and C1 f/f mice, in which CARMA1 was deleted in all (FcreERT2) or half (FcreERT2/+) of Treg cells. Arrow indicates tamoxifen treatment start. g, In situ expression in tumour tissue of effector cytokines by YFP+ Treg five days after CARMA1 deletion in half or all Treg cells. h, MHC-II expression on tumour-associated macrophages in D4M.3A-implanted Fcre/+ × C1+/+, C1f/+ or C1f/f mice. i, D4M.3A tumour growth in mice treated with depleting anti-CD8 antibody from day 8 and treated with mepazine or vehicle from day 9. j, D4M.3A tumour growth in Fcre/+ × C1+/+ or C1f/f mice treated with mepazine or vehicle starting on day 9. Data are mean and individual replicates or s.e.m. In a–c, g–i, P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. In d, *P < 0.05 versus C1+/+, &P < 0.05 versus C1+/+ + IKK2ca. In f, *P < 0.05 versus FcreERT2 × C1+/+, #P < 0.05 versus FcreERT2/+ × C1f/f. In j, *P < 0.05 versus C1+/+, #P < 0.05 versus C1+/+ + mepazine (one-way ANOVA with Tukey post hoc test in a–c, g, h; two-way ANOVA with Bonferroni post hoc test in d, f, j: two-tailed Student’s t-test in i).
Extended Data Fig. 10 Mepazine effects on the tumour microenvironment.
a, YFP+ Treg cells were sorted from Fcre × C1+/+ mice and treated with 10 μM mepazine or vehicle for 8 or 24 h with or without concurrent anti-CD3/28 monoclonal antibody TCR stimulation (8-h time point only). Expression of FOXP3, markers of eTreg cell differentiation, cell viability, and frequency of eTreg cells were recorded. b, c, RT–qPCR analysis of expression of Ifng and genes of adaptive immune resistance (the PD-L1 genes Cd274 and Socs1), antigen presentation (Tap1), IFNγ signalling (Stat1 and Irf1), T-cell recruitment (Cxcl10), M1 macrophage-activation (Nos2) (b) and of Foxp3 and various Treg-cell-associated genes (c) in whole tumour tissue lysate after three days of treatment with mepazine or vehicle control. d–h, Composition of the tumour tissue immune infiltrate and frequencies of CD45+ cells (e) and of various immune cell subsets (f) as well Ki67 expression by Tconv cells (g) and MHC-II expression by macrophages (h) after three days of treatment with mepazine or vehicle control. i, Effector cytokine co-expression by tumour-infiltrating Treg cells after 12 days of treatment with mepazine and anti-PD-1 antibody. j, Synergistic tumour control of MC38 colon carcinoma through anti-PD-1 and mepazine combination treatment in female C57BL/6 hosts. Numbers in parentheses indicate fraction of mice without relapse for more than 12 months after discontinuation of treatment. In j, *P < 0.05 versus vehicle, #P < 0.05 versus anti-PD-1, and &P < 0.05 versus mepazine. In all other panels, *P < 0.05, **P < 0.01, ***P < 0.001 (two-tailed Student’s t-test in a–c, e–h; one-way ANOVA with Tukey post hoc test in i; two-way ANOVA with Bonferroni post hoc test in j).
Supplementary information
Supplementary Information
This file contains Supplementary Materials and Methods: R scripts used in the analyses of RNA-sequencing data.
Supplementary Table
This file contains Supplementary Table 1: Sheets 1, 2: Differentially expressed genes (DEGs, defined by fold change > 2 and padj <0.05) in YFP+ cTreg (sheet 1) or YFP+ eTreg (sheet 2) from Foxp3Cre/+ x CARMA1flox/flow compared to Foxp3Cre x CARMA1+/+ mice. Sheet 3: Differences in eTreg signature genes between YFP+ eTreg from Foxp3Cre/+ x CARMA1flox/flox versus Foxp3Cre x CARMA1+/+ mice.
Rights and permissions
About this article
Cite this article
Di Pilato, M., Kim, E.Y., Cadilha, B.L. et al. Targeting the CBM complex causes Treg cells to prime tumours for immune checkpoint therapy. Nature 570, 112–116 (2019). https://doi.org/10.1038/s41586-019-1215-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1215-2
This article is cited by
-
Potential anti-tumor effects of regulatory T cells in the tumor microenvironment: a review
Journal of Translational Medicine (2024)
-
Therapeutic and immunomodulatory potentials of mesenchymal stromal/stem cells and immune checkpoints related molecules
Biomarker Research (2024)
-
Functional CRISPR screens in T cells reveal new opportunities for cancer immunotherapies
Molecular Cancer (2024)
-
IL-23 stabilizes an effector Treg cell program in the tumor microenvironment
Nature Immunology (2024)
-
Treg plasticity and human diseases
Inflammation Research (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.