Abstract
High-grade serous carcinoma has a poor prognosis, owing primarily to its early dissemination throughout the abdominal cavity. Genomic and proteomic approaches have provided snapshots of the proteogenomics of ovarian cancer1,2, but a systematic examination of both the tumour and stromal compartments is critical in understanding ovarian cancer metastasis. Here we develop a label-free proteomic workflow to analyse as few as 5,000 formalin-fixed, paraffin-embedded cells microdissected from each compartment. The tumour proteome was stable during progression from in situ lesions to metastatic disease; however, the metastasis-associated stroma was characterized by a highly conserved proteomic signature, prominently including the methyltransferase nicotinamide N-methyltransferase (NNMT) and several of the proteins that it regulates. Stromal NNMT expression was necessary and sufficient for functional aspects of the cancer-associated fibroblast (CAF) phenotype, including the expression of CAF markers and the secretion of cytokines and oncogenic extracellular matrix. Stromal NNMT expression supported ovarian cancer migration, proliferation and in vivo growth and metastasis. Expression of NNMT in CAFs led to depletion of S-adenosyl methionine and reduction in histone methylation associated with widespread gene expression changes in the tumour stroma. This work supports the use of ultra-low-input proteomics to identify candidate drivers of disease phenotypes. NNMT is a central, metabolic regulator of CAF differentiation and cancer progression in the stroma that may be therapeutically targeted.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data supporting the findings of this study are available within the paper, Extended Data Figs. 1–10, the Source Data (of Figs. 2–4 and Extended Data Figs. 2–10), publicly accessible repositories, or from the corresponding author upon reasonable request. Proteomic data are available online at http://maxqb.biochem.mpg.de/mxdb/project/show/9373012627500 and have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD006396) via the PRIDE partner repository with the dataset identifier PXD006396. Raw and processed data are available in the Gene Expression Omnibus (GEO) database (gene expression microarray data, GSE112497; DNA methylation array, GSE126672; RNA-sequencing data, GSE124014; ChIP-seq data, GSE124015). The MaxQuant quantitative proteomics software package and Perseus data analysis software environment are freely available: http://www.biochem.mpg.de/5111733/software. Supplementary Data are available online.
References
Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011).
Zhang, H. et al. Integrated proteogenomic characterization of human high-grade serous ovarian cancer. Cell 166, 755–765 (2016).
Kuhn, E. et al. TP53 mutations in serous tubal intraepithelial carcinoma and concurrent pelvic high-grade serous carcinoma-evidence supporting the clonal relationship of the two lesions. J. Pathol. 226, 421–426 (2012).
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Francavilla, C. et al. Phosphoproteomics of primary cells reveals druggable kinase signatures in ovarian cancer. Cell Reports 18, 3242–3256 (2017).
Coscia, F. et al. Integrative proteomic profiling of ovarian cancer cell lines reveals precursor cell associated proteins and functional status. Nat. Commun. 7, 12645 (2016).
Polyak, K., Haviv, I. & Campbell, I. G. Co-evolution of tumor cells and their microenvironment. Trends Genet. 25, 30–38 (2009).
Kalluri, R. The biology and function of fibroblasts in cancer. Nat. Rev. Cancer 16, 582–598 (2016).
Aebersold, R. & Mann, M. Mass-spectrometric exploration of proteome structure and function. Nature 537, 347–355 (2016).
Altelaar, A. F. & Heck, A. J. Trends in ultrasensitive proteomics. Curr. Opin. Chem. Biol. 16, 206–213 (2012).
Nieman, K. M. et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat. Med. 17, 1498–1503 (2011).
Pissios, P. Nicotinamide N-methyltransferase: more than a vitamin B3 clearance enzyme. Trends Endocrinol. Metab. 28, 340–353 (2017).
Aksoy, S., Szumlanski, C. L. & Weinshilboum, R. M. Human liver nicotinamide N-methyltransferase. cDNA cloning, expression, and biochemical characterization. J. Biol. Chem. 269, 14835–14840 (1994).
Ulanovskaya, O. A., Zuhl, A. M. & Cravatt, B. F. NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat. Chem. Biol. 9, 300–306 (2013).
Kraus, D. et al. Nicotinamide N-methyltransferase knockdown protects against diet-induced obesity. Nature 508, 258–262 (2014).
Hong, S. et al. Nicotinamide N-methyltransferase regulates hepatic nutrient metabolism through Sirt1 protein stabilization. Nat. Med. 21, 887–894 (2015).
Sperber, H. et al. The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nat. Cell Biol. 17, 1523–1535 (2015).
Perets, R. et al. Transformation of the fallopian tube secretory epithelium leads to high-grade serous ovarian cancer in Brca;Tp53;Pten models. Cancer Cell 24, 751–765 (2013).
Orimo, A. et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell 121, 335–348 (2005).
Olumi, A. F. et al. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res. 59, 5002–5011 (1999).
Neelakantan, H. et al. Selective and membrane-permeable small molecule inhibitors of nicotinamide N-methyltransferase reverse high fat diet-induced obesity in mice. Biochem. Pharmacol. 147, 141–152 (2018).
Ferro, A. J., Vandenbark, A. A. & MacDonald, M. R. Inactivation of S-adenosylhomocysteine hydrolase by 5′-deoxy-5′-methylthioadenosine. Biochem. Biophys. Res. Commun. 100, 523–531 (1981).
Zheng, Y. et al. Total kinetic analysis reveals how combinatorial methylation patterns are established on lysines 27 and 36 of histone H3. Proc. Natl Acad. Sci. USA 109, 13549–13554 (2012).
Leung, C. S. et al. Calcium-dependent FAK/CREB/TNNC1 signalling mediates the effect of stromal MFAP5 on ovarian cancer metastatic potential. Nat. Commun. 5, 5092 (2014).
Kenny, H. A. et al. Targeting the urokinase plasminogen activator receptor inhibits ovarian cancer metastasis. Clin. Cancer Res. 17, 459–471 (2011).
Pozniak, Y. et al. System-wide clinical proteomics of breast cancer reveals global remodeling of tissue homeostasis. Cell Syst. 2, 172–184 (2016).
Eckert, M. A. et al. Genomics of ovarian cancer progression reveals diverse metastatic trajectories including intraepithelial metastasis to the fallopian tube. Cancer Discov. 6, 1342–1351 (2016).
Labidi-Galy, S. I. et al. High grade serous ovarian carcinomas originate in the fallopian tube. Nat. Commun. 8, 1093 (2017).
Hu, M. et al. Distinct epigenetic changes in the stromal cells of breast cancers. Nat. Genet. 37, 899–905 (2005).
Zong, Y. et al. Stromal epigenetic dysregulation is sufficient to initiate mouse prostate cancer via paracrine Wnt signaling. Proc. Natl Acad. Sci. USA 109, E3395–E3404 (2012).
Wang, H. et al. Development and evaluation of a micro- and nanoscale proteomic sample preparation method. J. Proteome Res. 4, 2397–2403 (2005).
Roulhac, P. L. et al. Microproteomics: quantitative proteomic profiling of small numbers of laser-captured cells. Cold Spring Harb. Protoc. 2011, prot5573 (2011).
Scheltema, R. A. et al. The Q Exactive HF, a Benchtop mass spectrometer with a pre-filter, high-performance quadrupole and an ultra-high-field Orbitrap analyzer. Mol. Cell. Proteomics 13, 3698–3708 (2014).
Michalski, A. et al. Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol. Cell. Proteomics 10, M111 (2011).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Cox, J. et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 (2011).
Cox, J. et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 (2014).
Tyanova, S. et al. Proteomic maps of breast cancer subtypes. Nat. Commun. 7, 10259 (2016).
Tusher, V. G., Tibshirani, R. & Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA 98, 5116–5121 (2001).
Kenny, H. A. et al. Mesothelial cells promote early ovarian cancer metastasis through fibronectin secretion. J. Clin. Invest. 124, 4614–4628 (2014).
Mitra, A. K. et al. MicroRNAs reprogram normal fibroblasts into cancer-associated fibroblasts in ovarian cancer. Cancer Discov. 2, 1100–1108 (2012).
Chang, J. W., Lee, G., Coukos, J. S. & Moellering, R. E. Profiling Reactive Metabolites via Chemical Trapping and Targeted Mass Spectrometry. Anal. Chem. 88, 6658–6661 (2016).
Cajka, T. & Fiehn, O. LC-MS-based lipidomics and automated identification of lipids using the LipidBlast in-silico MS/MS library. Methods Mol. Biol. 1609, 149–170 (2017).
Cajka, T., Smilowitz, J. T. & Fiehn, O. Validating quantitative untargeted lipidomics across nine liquid chromatography-high-resolution mass spectrometry platforms. Anal. Chem. 89, 12360–12368 (2017).
Showalter, M. R. et al. Obesogenic diets alter metabolism in mice. PLoS ONE 13, e0190632 (2018).
Tu, L. N. et al. Metabolomic characteristics of cholesterol-induced non-obese nonalcoholic fatty liver disease in mice. Sci. Rep. 7, 6120 (2017).
Fahrmann, J. et al. Systemic alterations in the metabolome of diabetic NOD mice delineate increased oxidative stress accompanied by reduced inflammation and hypertriglyceremia. Am. J. Physiol. Endocrinol. Metab. 308, E978–E989 (2015).
Fiehn, O. Metabolomics by gas chromatography-mass spectrometry: combined targeted and untargeted profiling. Curr. Protoc. Mol. Biol. 114, 1–32, 32 (2016).
Skogerson, K., Wohlgemuth, G., Barupal, D. K. & Fiehn, O. The volatile compound BinBase mass spectral database. BMC Bioinformatics 12, 321 (2011).
Sumner, L. W. et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3, 211–221 (2007).
Pidsley, R. et al. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 17, 208 (2016).
Allhoff, M., Seré, K., F Pires, J., Zenke, M. & G Costa, I. Differential peak calling of ChIP-seq signals with replicates with THOR. Nucleic Acids Res. 44, e153 (2016).
Acknowledgements
We thank H. A. Kenny, K. Watters and A. Mukherjee from the University of Chicago ovarian cancer laboratory for helpful discussions; and G. Isenberg, University of Chicago, for editing the manuscript. This work was supported by a Marsha Rivkin Foundation award (M.A.E.), National Cancer Institute (NCI) grants CA111882 and CA211916 (E.L.), the Ludwig Institute for Cancer Research (E.L.), the Arthur L. and Lee G. Herbst Professorship (E.L.), funding support from S. and J. Harris, M. Field, J. Kane and A. Gerry (M.A.E. and S.D.Y.), NIH grant CA175399 and DP2GM128199 (R.E.M.), V Foundation for Cancer Research (V2016-020 to R.E.M.), the Körber Foundation/Körber European Science Prize (M.M.), the Max-Planck Society for the Advancement of Science (M.M.), the Novo Nordisk Foundation (grant agreement NNF14CC0001 and NNF15CC0001; F.C. and M.M.), and University of Chicago Cancer Center Support Grant P30CA014599.
Reviewer information
Nature thanks Amina Qutub and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
The study was conceived by M.A.E. and E.L. Proteomic sample preparation, analysis and interpretation were performed by F.C. under supervision of M.M. Experiments were designed by M.A.E., F.C., R.E.M., M.M. and E.L. Tissues were microdissected by M.A.E. and M.C. Experiments with primary and cancer cells were performed by M.A.E., S.P., A.C., D.A.N. and S.M.T. Animal experiments were performed by M.A.E., S.P., S.M.T. and A.C. Targeted metabolite analyses and inhibitor synthesis were performed by J.W.C and G.L. under the supervision of R.E.M. Data were analysed by M.A.E., F.C. and J.W.C. Human tissues for isolation of primary cells and proteomic analysis were identified and collected by S.D.Y. and E.L. Global metabolomics were performed, analysed and interpreted by I.B. under the supervision of O.F. Bioinformatics analyses of sequencing data were performed by K.M.H. under the supervision of J.A. Autochthonous mice were maintained and tissues collected and provided by R.P. Tissue microarrays were interpreted and scored by S.M.M. and R.R.L. Figures were prepared by M.A.E., F.C. and K.M.H. The paper was written by M.A.E. and E.L. The paper was edited by M.A.E., F.C., M.M., R.E.M. and E.L. All authors reviewed and provided feedback on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Quantitative proteomics of low-input samples.
a, MaxLFQ label-free quantitation values and dynamic range are similar across all anatomic sites and in both tumour (left) and stroma (right) samples. b, Example of experimental replicates of microdissection, protein extraction, and quantitative proteomics with Pearson correlation of 0.98. n = 2 technical replicates. c, Unsupervised hierarchical clustering of all proteomic samples leads to clustering of tumour (n = 43) and stromal (n = 42) samples characterized by proteomic signatures associated with the indicated pathways (green and purple boxes, respectively). Enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways for tumour (left) and stroma (right) clusters are annotated based on a Fisher’s exact test (FDR < 0.02). d, One-dimensional principal component analysis of all tumour (n = 43) and stromal (n = 42) samples. Component 1 accounts for 23.5% of the total data variation. e, Volcano plot comparing all tumour and all stromal samples reveals enrichment of known markers characterizing tumour (green; n = 43 samples) and stromal (purple; n = 42 samples) components of the tumour (MUC16 is CA-125). Two-sided t-test FDR < 0.01, n = 11 patients.
Extended Data Fig. 2 HGSC progression is characterized by patient-specific signatures in the tumour compartment and anatomic site-specific signatures in the stroma.
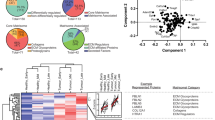
a, FABP4 expression in tumour (n = 36) and stromal (n = 38) compartments derived from label-free, quantitative proteomics. One-way ANOVA. b, Plot of ANOVA P value of tumour (left; n = 43) or stroma (right; n = 42) samples calculated by patient (y axis) or anatomic site (x axis). In the tumour compartment, differential expression of proteins is driven by the patient grouping; in the stromal compartment, anatomic site-specific differences are more pertinent. c, Principal component analysis resolves an omental stromal cluster (n = 42). No anatomic-site-specific clusters are present in tumour samples (n = 43). d, Left and right panels show proportions of all proteins that are significantly different by patient (red) or anatomic site/compartment (blue). One thousand, four hundred and seventy-four proteins are differentially expressed in the tumour compartment between patients whereas only 30 stromal proteins are significantly different between patients. In respect to the compartment (tumour/stroma, blue; grey is undetected) one protein is differentially expressed in the tumour compartment (FABP4), whereas 128 proteins are differentially expressed in the stroma. e, Unsupervised hierarchical clustering of tumour (left, 1,474 significant proteins from d) and stroma (right, 128 significant proteins from d) proteins reveals patient-specific clustering in the tumour compartment (n = 43) whereas the stromal samples (n = 42) cluster by anatomic site. Note, all omental samples (red) across all patients cluster together. f, Unsupervised hierarchical clustering of only stromal proteins (n = 42) that are differentially expressed between primary sites (FT and Ov) and metastases (Om) reveal anatomic site-specific clusters, including a core signature of 21 proteins consistently upregulated in the stroma of omental metastases (box). g, Expression of the 21-protein signature in the TCGA subtypes reveals enhanced expression in mesenchymal subtype (n = 21 proteins).
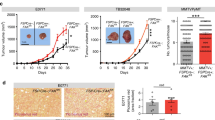
Extended Data Fig. 3 NNMT is highly expressed in the stroma of ovarian cancers.
a, Quantitative proteomics of the stroma finds increased expression of NNMT in omental metastases (n = 11) compared to normal (normal fallopian tube (nFT; n = 5), normal ovarian (nOv; n = 5), and normal omentum (nOm; n = 6) and primary tumour tissues (STIC, n = 9; invasive fallopian tube (FT), n = 10; and invasive ovarian (Ov), n = 11). b, Human omental metastasis tissue stained with NNMT-specific antibody or IgG isotype control; no non-specific staining is observed. Scale bar, 50 μm. c, Representative NNMT IHC of peritoneal metastasis. d, NNMT IHC of an omentum with micrometastases (1) and larger metastases (2 and 3). NNMT is detected in the stroma of very early metastases. e, Quantification of tumour NNMT staining in tissue microarray analysis. Chi-squared test, n = 169 ovarian, 135 omental, and 92 peritoneal samples. f, Representative IHC of NNMT in the stroma of metastases in an autochthonous model of ovarian cancer (top, Pax8:Trp53mutPten−/−Brca1mut) and a syngeneic model (bottom, ID8 intraperitoneal xenograft). g, NNMT is expressed in the stroma of breast and colon cancers, but not normal breast stroma.
Extended Data Fig. 4 NNMT promotes acquisition and maintenance of the CAF phenotype.
a, RT–qPCR for NNMT in CAFs expressing an shRNA against NNMT. Two-sided t-test, n = 3 biological replicates. b, Immunofluorescence analysis of SMA reveals a decrease of SMA stress fibres upon knockdown of NNMT in CAFs. c, Representative phase contrast images of normal omental fibroblasts and primary CAFs expressing shCtrl and shNNMT constructs. Upon knockdown of NNMT a reversion of CAF morphology to more closely resemble normal omental fibroblasts is observed. Scale bar, 50 μm. d, e, Relative mRNA (two-sided t-test, n = 3 biological replicates) (d) and immunoblotting of fibronectin and SMA in CAFs transfected with the indicated siRNAs (e). f, Knockdown of NNMT in stromal cells attenuates expression of fibronectin, SMA and SNAI1 following TGF-β treatment. g, Cytokine array. A shNNMT construct was expressed in primary human CAFs and a cytokine array performed on the conditioned medium. Cytokines downregulated upon knockdown of NNMT are highlighted in blue, those increased in red. Individual cytokines are indicated by Roman numerals. n = 2 technical replicates. h–k, GSEA of genes regulated by NNMT in stromal cells reveals associations with the epithelial–mesenchymal transition hallmark gene set (h–i); proteins enriched in the metastatic stroma compared to primary tumour stroma (proteomics-our data) (j); and the TCGA mesenchymal signature (RNA) (k). l, Representative images of chemotaxis of indicated HGSC cells in response to conditioned medium from CAFs expressing shCtrl or shNNMT constructs. m, Proliferation of HeyA8 cells grown in direct co-culture with CAFs transfected with the indicated siRNAs. Two-sided t-test, n = 3 biological replicates. All bar graphs represent mean of data and error bars are s.e.m.
Extended Data Fig. 5 NNMT regulates nicotinamide metabolism and sirtuins.
a, NNMT depletes nicotinamide (NA), a precursor to NAD+ biosynthesis; NAD+ is an essential cofactor for sirtuin deacetylase activity which broadly regulates acetylation of proteins including tubulin and histones. NamPRT: nicotinamide phosphoribosyl transferase; NMNAT: nicotinamide mononucleotide adenylyltransferase; NADK: nicotinamide adenine dinucleotide kinase. b, Quantification of NAD+, NADH, and the NAD+–NADH ratio in CAFs expressing shCtrl or shNNMT constructs. Two-sided t-test, n = 3 biological replicates. c, Relative expression of genes regulated by sirtuin activity in CAFs expressing shCtrl or shNNMT constructs. Two-sided t-test, n = 3 biological replicates. Catalase (CAT), succinate dehydrogenase subunit B (SDHB). d, Acetylation of α-tubulin and histone H3K9 are reduced upon knockdown of NNMT in CAFs. e, Quantification of α-tubulin acetylation immunoblots in CAFs expressing the indicated constructs. Two-sided t-test, n = 3 biological replicates. All bar graphs represent mean of data and error bars are s.e.m.
Extended Data Fig. 6 NNMT regulates global metabolism.
a, Untargeted metabolite profiling reveals that NNMT induces conserved metabolic changes. Metabolites that are significantly changed upon both knockdown and overexpression of NNMT are shown (P < 0.05). Samples cluster by NNMT expression status rather than cell of origin. b, The polyamine pathway consumes decarboxylated SAM (AdoMetDC, generated from SAM by the action of SAM decarboxylase; SAMDC) and generates 5-methylthioadenosine (5-MTA) during the conversion of putrescine to spermidine and spermine. ODC, ornithine decarboxylase; SRM, spermidine synthase; SMS, spermine synthase. c, d, 5-MTA and putrescine levels are positively associated with NNMT expression in both fibroblasts and CAFs. All bar graphs represent mean of data and error bars are s.e.m.
Extended Data Fig. 7 NNMT regulates DNA and histone methylation in the stroma.
a, Overexpression of NNMT leads to hypomethylation. Distribution of significantly different β-values (Benjamini–Hochberg adjusted P value < 0.01) within 1,500 bp of TSSs in fibroblasts expressing the indicated constructs as assessed by global DNA methylation arrays. β-values are lower in cells overexpressing NNMT, indicating decreased DNA methylation associated with TSSs. n = 2 biological replicates. b, KEGG pathway analysis of genes differentially methylated upon both NNMT overexpression and knockdown in normal fibroblasts and CAFs, respectively. FDR (B&H), Benjamini–Hochberg P value. c, Treatment of normal fibroblasts with the DNA methylation inhibitor 5-azacytidine (5-Aza) (100 nM) leads to an increase in SMA expression. d, Collagen contractility of normal fibroblasts is increased following treatment with 5-azacytidine (100 nM). Two-sided t-test, n = 3 biological replicates. e, NNMT-mediated attenuation of H3K4 trimethylation is metabolically regulated and can be rescued by high (200 µM) methionine concentrations. f, Quantification of H3K4 trimethylation immunoblotting in cells cultured with medium containing 10 or 200 μM methionine. Two-sided t-test, n = 3 biological replicates. All bar graphs represent mean of data and error bars are s.e.m.
Extended Data Fig. 8 NNMT regulates stromal histone methylation.
a, Genes regulated by the downregulation of H3K27 trimethylation (Acevedo gene set from Molecule Signatures Database; software.broadinstitute.org/gsea/msigdb) are enriched in genes regulated by NNMT overexpression as assessed by GSEA. b, Overexpression of NNMT in normal fibroblasts followed by H3K27me3 immunoprecipitation and ChIP-seq. Violin plot of ChIP-seq peak size distribution corresponding to H3K27me3 occupancy at the promoter of genes significantly upregulated (gene expression FDR P < 0.05 from RNA-sequencing analysis) in NNMT overexpressing fibroblasts; NNMT overexpression leads to a significant reduction in H3K27me3 occupancy. Solid black line indicates median, dashed lines the quartiles. c, Same experimental setup as in b. H3K27me3 ChIP-seq of normal fibroblasts overexpressing NNMT. H3K27me3 read density plots (top) and heat maps (bottom) in relationship to genes downregulated, upregulated or unchanged (gene expression FDR P < 0.05 from RNA sequencing analysis) in response to NNMT overexpression. ChIP-seq data are binned into expression groups from parallel RNA sequencing analysis. n = 3 biological replicates. d, Overexpression of NNMT leads to genome-wide H3K27 hypomethylation (reduced H3K27me3 occupancy) at gene promoters adjacent to TSSs. Thus, NNMT expression upregulates expression of a significant number of genes by relieving H3K27me3-mediated transcriptional suppression. e, Immunohistochemistry of COMP in an omental metastasis. Scale bar, 100 μm. f, COMP gene expression in CAFs or fibroblasts expressing the indicated constructs. Two-sided t-test, n = 3 biological replicates. g, Coverage tracks of H3K27me3 ChIP-seq analysis of fibroblasts expressing control or NNMT overexpression constructs. Genes regulated by NNMT expression (Comp) display reduced coverage depth while non-perturbed loci (Hoxa10/Hoxa11) are sequenced to similar depths. Comp, Hoxa10 and Hoxa11 exon/intro structure are indicated and Comp gene direction and TSS. All bar graphs represent mean of data and error bars are s.e.m.
Extended Data Fig. 9 Inhibition of H3K27 trimethylation restores features of the CAF phenotype.
a, Expression of CAF marker genes in CAFs expressing shNNMT and treated with the EZH2 histone methyltransferase inhibitor DZNep. Two-sided t-test, n = 3 biological replicates. b, Immunoblot of CAFs expressing a shNNMT construct treated with the general methyltransferase inhibitor 3-DZA. c, d, Knockdown of EZH2 in CAFs expressing a shNNMT construct leads to decreased H3K27 trimethylation (c) and increased collagen contractility (d). Two-sided t-test, n = 3 biological replicates. All bar graphs represent mean of data and error bars are s.e.m.
Extended Data Fig. 10 NNMT inhibition.
a, Raw tumour luminescence values of HeyA8/CAF subcutaneous xenograft at 7 and 14 days following implantation. CAF expressing shNNMT construct (or control; shNNMT). b, In vitro NNMT enzymatic activity assay in response to NNMTi treatment, n = 3 experimental replicates. c, Cellular thermal shift assay performed with CAOV3 cells at the indicated temperatures to establish aggregation temperature for NNMT. d, Cellular thermal shift assay suggests cellular target engagement of NNMTi at micromolar concentrations. e, Small molecule inhibition of NNMT in CAFs leads to reduction in tubulin acetylation. f, Quantification of H3K4me3 immunoblotting in fibroblasts expressing the indicated constructs following treatment with NNMTi; H3K4 trimethylation is only increased in cells expressing NNMT after treatment with the inhibitor, indicating that effects on histone methylation are NNMT-specific. Two-sided t-test, n = 3 biological replicates. g, Immunoblotting: NNMT is only expressed in the CAOV3 ovarian cancer cell line. h, i, Treatment of ovarian cancer cells (h) or CAFs (i) with the NNMTi only reduced cell viability at high concentrations, n = 3 biological replicates. j, Representative images and quantification of cancer cell proliferation in mouse tumours treated with NNMTi or vehicle control. Two-sided t-test, n = 8 biological replicates. Scale bar, 100 μm. k, Mouse omental tumour tissue stained with H3K27me3 or IgG isotype control antibody demonstrates specificity of antibody. Scale bar, 10 μm. l, Immunofluorescence of H3K27me3 in omental metastases from mice in Fig. 4d confirms that NNMT inhibition increases histone methylation in the tumour microenvironment. Stromal compartment is highlighted by dashed lines. Scale bar, 50 μm. m, Quantification of H3K27me3 nuclear staining intensity of stroma or tumour compartment in omental tumours from mice treated with vehicle or NNMTi. H3K27 trimethylation is increased in the stromal compartment. Two-sided t-test, n = 8 biological replicates. n, Representative images of ovarian tumours expressing stromal NNMT. o, Kaplan–Meier curve of recurrence-free survival of patients with low (black) or high (red) stromal NNMT expression in primary tumour sites (two-tailed test). p, q, Kaplan–Meier curve of overall (p) and recurrence-free survival (q) of patients with low (black) or high (red) tumour NNMT expression in primary sites (two-tailed test). All bar graphs represent mean of data and error bars are s.e.m.
Supplementary information
Supplementary Information
This file contains the uncropped immunoblots for all Figures and Extended Data Figures and a Supplementary Table Guide.
Supplementary Tables
This file contains Supplementary Tables 1-14 – see Supplementary Information document for a full table guide.
Source data
Rights and permissions
About this article
Cite this article
Eckert, M.A., Coscia, F., Chryplewicz, A. et al. Proteomics reveals NNMT as a master metabolic regulator of cancer-associated fibroblasts. Nature 569, 723–728 (2019). https://doi.org/10.1038/s41586-019-1173-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1173-8
This article is cited by
-
Cancer-associated fibroblast-secreted FGF7 as an ovarian cancer progression promoter
Journal of Translational Medicine (2024)
-
The miR-1290/OGN axis in ovarian cancer-associated fibroblasts modulates cancer cell proliferation and invasion
Journal of Ovarian Research (2024)
-
Quantitative proteomic analysis of HER2 protein expression in PDAC tumors
Clinical Proteomics (2024)
-
Stromal cartilage oligomeric matrix protein as a tumorigenic driver in ovarian cancer via Notch3 signaling and epithelial-to-mesenchymal transition
Journal of Translational Medicine (2024)
-
Metabolomic machine learning predictor for diagnosis and prognosis of gastric cancer
Nature Communications (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.