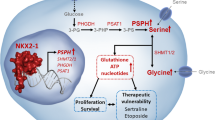
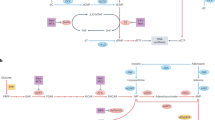
Abstract
Precision oncology hinges on linking tumour genotype with molecularly targeted drugs1; however, targeting the frequently dysregulated metabolic landscape of cancer has proven to be a major challenge2. Here we show that tissue context is the major determinant of dependence on the nicotinamide adenine dinucleotide (NAD) metabolic pathway in cancer. By analysing more than 7,000 tumours and 2,600 matched normal samples of 19 tissue types, coupled with mathematical modelling and extensive in vitro and in vivo analyses, we identify a simple and actionable set of ‘rules’. If the rate-limiting enzyme of de novo NAD synthesis, NAPRT, is highly expressed in a normal tissue type, cancers that arise from that tissue will have a high frequency of NAPRT amplification and be completely and irreversibly dependent on NAPRT for survival. By contrast, tumours that arise from normal tissues that do not express NAPRT highly are entirely dependent on the NAD salvage pathway for survival. We identify the previously unknown enhancer that underlies this dependence. Amplification of NAPRT is shown to generate a pharmacologically actionable tumour cell dependence for survival. Dependence on another rate-limiting enzyme of the NAD synthesis pathway, NAMPT, as a result of enhancer remodelling is subject to resistance by NMRK1-dependent synthesis of NAD. These results identify a central role for tissue context in determining the choice of NAD biosynthetic pathway, explain the failure of NAMPT inhibitors, and pave the way for more effective treatments.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data supporting the findings of this study are available within the paper and its Supplementary Information. Source Data for all figures are available online. Data are available from the corresponding author upon reasonable request.
References
Roychowdhury, S. & Chinnaiyan, A. M. Translating genomics for precision cancer medicine. Annu. Rev. Genomics Hum. Genet. 15, 395–415 (2014).
Mayers, J. R. & Vander Heiden, M. G. Nature and nurture: what determines tumor metabolic phenotypes? Cancer Res. 77, 3131–3134 (2017).
Cantó, C., Menzies, K. J. & Auwerx, J. NAD+ metabolism and the control of energy homeostasis: a balancing act between mitochondria and the nucleus. Cell Metab. 22, 31–53 (2015).
Verdin, E. NAD+ in aging, metabolism, and neurodegeneration. Science 350, 1208–1213 (2015).
Vander Heiden, M. G. & DeBerardinis, R. J. Understanding the intersections between metabolism and cancer biology. Cell 168, 657–669 (2017).
Pavlova, N. N. & Thompson, C. B. The Emerging hallmarks of cancer metabolism. Cell Metab. 23, 27–47 (2016).
Liu, L. et al. Quantitative analysis of NAD synthesis-breakdown fluxes. Cell Metab. 27, 1067–1080 (2018).
Chiarugi, A., Dölle, C., Felici, R. & Ziegler, M. The NAD metabolome—a key determinant of cancer cell biology. Nat. Rev. Cancer 12, 741–752 (2012).
Ryu, K. W. et al. Metabolic regulation of transcription through compartmentalized NAD+ biosynthesis. Science 360, eaan5780 (2018).
Kaelin, W. G. Jr & McKnight, S. L. Influence of metabolism on epigenetics and disease. Cell 153, 56–69 (2013).
Bogan, K. L. & Brenner, C. Nicotinic acid, nicotinamide, and nicotinamide riboside: a molecular evaluation of NAD+ precursor vitamins in human nutrition. Annu. Rev. Nutr. 28, 115–130 (2008).
Katsyuba E. et al. De novo NAD+ synthesis enhances mitochondrial function and improves health. Nature 563, 354–359 (2018).
Tateishi, K. et al. Extreme vulnerability of IDH1 mutant cancers to NAD+ depletion. Cancer Cell 28, 773–784 (2015).
Piacente, F. et al. Nicotinic acid phosphoribosyltransferase regulates cancer cell metabolism, susceptibility to NAMPT inhibitors, and DNA repair. Cancer Res. 77, 3857–3869 (2017).
Hnisz, D. et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013).
Leung, D. et al. Integrative analysis of haplotype-resolved epigenomes across human tissues. Nature 518, 350–354 (2015).
Gilbert, L. A. et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013).
Frederick, D. W. et al. Loss of NAD homeostasis leads to progressive and reversible degeneration of skeletal muscle. Cell Metab. 24, 269–282 (2016).
Moro, W. B., Yang, Z., Kane, T. A., Brouillette, C. G. & Brouillette, W. J. Virtual screening to identify lead inhibitors for bacterial NAD synthetase (NADs). Bioorg. Med. Chem. Lett. 19, 2001–2005 (2009).
Devedjiev, Y. et al. Stabilization of active-site loops in NH3-dependent NAD+ synthetase from Bacillus subtilis. Acta Crystallogr. D 57, 806–812 (2001).
Hara, N. et al. Molecular identification of human glutamine- and ammonia-dependent NAD synthetases. Carbon-nitrogen hydrolase domain confers glutamine dependency. J. Biol. Chem. 278, 10914–10921 (2003).
Velu, S. E., Luan, C. H., Delucas, L. J., Brouillette, C. G. & Brouillette, W. J. Tethered dimer inhibitors of NAD synthetase: parallel synthesis of an aryl-substituted SAR library. J. Comb. Chem. 7, 898–904 (2005).
Wang, X. et al. Design, synthesis, and evaluation of substituted nicotinamide adenine dinucleotide (NAD+) synthetase inhibitors as potential antitubercular agents. Bioorg. Med. Chem. Lett. 27, 4426–4430 (2017).
Heintzman, N. D. et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311–318 (2007).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Lonsdale, J. et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genetics 45, 580–585 (2013).
Grossman, R. L. et al. Toward a shared vision for cancer genomic data. N. Engl. J. Med. 375, 1109–1112 (2016).
Hartigan, J. A. & Hartigan, P. M. The dip test of unimodality. Ann. Stat. 13, 70–84 (1985).
Hasmann, M. & Schemainda, I. FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer Res. 63, 7436–7442 (2003).
Turner, K. M. et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature 543, 122–125 (2017).
Wojcik, M., Seidle, H. F., Bieganowski, P. & Brenner, C. Glutamine-dependent NAD+ synthetase. How a two-domain, three-substrate enzyme avoids waste. J. Biol. Chem. 281, 33395–33402 (2006).
Zalkin, H. NAD synthetase. Methods Enzymol. 113, 297–302 (1985).
Zhang, X. et al. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat. Genet. 48, 176–182 (2016).
Acknowledgements
We thank M. Meyerson for CRISPR interference plasmids; Mischel laboratory members and A. Shiau for suggestions; and A. Hwang for generating Extended Data Fig. 3a. This work was supported by Ludwig Institute for Cancer Research (P.S.M., B.R., F.F.), Defeat GBM program of the National Brain Tumor Society (P.S.M., F.F.), a Sharpe-National Brain Tumor Society Research Award (P.S.M.), NVIDIA Foundation, (P.S.M.), the Ben and Catherine Ivy foundation (P.S.M.), the Ziering Family Foundation in memory of Sigi Ziering (P.S.M.) and NIH grants T32 CA009253 (R.R.), CA121938 (E.B.), NS73831 (P.S.M.), GM114362 (V.B.), NS80939 (F.F.), and NSF grants NSF-IIS-1318386 and NSF-DB1-1458557 (V.B.).
Author information
Authors and Affiliations
Contributions
S.C., V.B., B.R. and P.S.M. conceived and designed the study and interpreted results. S.C., V.B., B.R., F.F. and P.S.M. wrote the manuscript with critical suggestions from all authors. S.C., C.Z., T.K., H.Y., K.T., U.R., Y.D., R.R., F.L., E.B. and J.B. conducted all experiments. C.Z. and U.R. contributed equally.
Corresponding author
Ethics declarations
Competing interests
P.S.M. and V.B. are co-founders of Pretzel Therapeutics, Inc., have equity in the company and serve as consultants. B.R. is a co-founder of Arima Genomics, Inc., and has equity in the company.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Tissue lineage-dependent, PH-pathway gene amplification in cancer.
a, Heatmap illustrating copy number (CN) alterations (z score) for NAMPT, NAPRT and NADSYN1 across cancer cell types (n = 54 cell types). b, Representative FISH images of cells in metaphase from two independent experiments with similar observations displaying NAPRT and NADSYN1 gene amplification on homogenously staining regions in PH-amplified (OV4PH-amp and KYSE510PH-amp) and non-PH-amplified (H460non-PH-amp) cancer cell lines as indicated. c, Violin plots of NAPRT (left) or NADSYN1 (right) mRNA expression against putative copy number alterations from several tumour types from CCLE (n = 947 from biologically independent samples including shallow/deep deletion). d, Violin plots of NAPRT or NADSYN1 mRNA expression stratified by NAPRT and NADSYN1 copy number alterations in multiple tumour types from cBioportal (ovarian adenocarcinoma: n = 403; oesophageal carcinoma: n = 150; hepatocellular carcinoma: n = 341; metastatic prostate adenocarcinoma: n = 99; breast carcinoma: n = 311; lung squamous cell carcinoma: n = 163; head and neck adenocarcinamoma: n = 503, from biologically independent samples). Violin plots display median, first and third quartiles. e, Heatmap illustrating differential gene expression profiles of NAD biosynthesis enzymes in PH-amplified and non-PH-amplified cancer cell types (z score, n = 54). Cancer cell lines amplified for the PH-pathway enzymes (NAPRT or NADSYN1) are denoted as ‘PH-amp’ in red, and cancer cell lines that are not amplified for NAPRT or NADSYN1 are denoted as ‘non-PH-amp’ in blue. f, Box and whisker plots showing normalized NAPRT transcript level (RPKM) in 19 distinct normal tissue of origin obtained from the GTEx and TCGA portal (www.gtexportal.org; www.portal.gdc.cancer.gov/repository). Box plot as in Fig. 1d, with all points are plotted according to the Tukey method. g, Bimodal distribution based on dip test of unimodality of two distributions stratified as ‘high’ and ‘low’ (n = 2,644 biologically independent samples). To classify tissues as having ‘high’ or ‘low’ gene expression, the critical point of distribution was chosen at 10 RPKM, at which the two distributions have identical density. h, Pearson correlation between expression of the NAPRT transcript (RPKM, z score) in 19 normal tissues and NAPRT or NADSYN1 copy number in 23 cancer types (n = 2,644 biologically independent samples). Statistical significance for mRNA expression against putative copy number alterations was assessed using two-tailed unpaired Student’s t-test (c, d). Experiments in a and e were repeated twice.
Extended Data Fig. 2 Non-cancer cells are not dependent on a single NAD biosynthetic pathway for survival.
a, Intracellular measurement of NAD+ levels in non-cancer cells after treatment with increasing doses of the NAMPT inhibitor FK-866 for 72 h. b, Representative images of non-cancer cells from one of two independent experiments, treated with increasing doses of FK-866 for 72 h. Both biological replicates showed similar results. Original magnification, ×10. c–f, Non-cancer cells transfected with siRNAs targeting NAMPT (siNAMPT), NMRK1 (siNMRK1) and NAPRT (siNAPRT), either individually or in combination. A non-targeting control siRNA (siNTC) was used a negative control. c, Scattered data plots with bars representing percentage of cell death as assessed by trypan blue exclusion assay in non-cancer cells. d, Intracellular measurement of NAD+ levels in non-cancer cells. e, Representative images of non-cancer cells from one of two independent experiments, transfected with siRNAs targeting NAMPT, NMRK1 and NAPRT, either individually or in combination. Both biological replicates showed similar results. Original magnification, ×10. f, Immunoblotting for cleaved caspase-3 (Cl-cas3) as a measure of cell death and to test for abundance of NAMPT, NMRK1 and NAPRT protein expression. Protein lysates from H460 cancer cells treated with etoposide (Etop) were used as a control, when immunoblotting for cleaved caspase-3. Actin was used as a loading control. Representative blots are from one of two independent experiments. Both biological replicates showed similar results. g, Intracellular measurement of NAD+ levels in non-cancer cells supplemented with exogenous NAD+ (200 µM) or with the indicated precursors, nicotinic acid (NA), nicotinamide (NM) or nicotinamide riboside (NR) at a concentration of 500 µM. Data are representative of five biological replicates from five independent experiments (a–d, g). Data are mean ± s.d. P values determined by one-way ANOVA with Tukey’s multiple comparisons test. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 3 Tissue context determines the NAD metabolic pathway dependence of cancer cells.
a, Schematic overview of 54 distinct established cancer models of 13 histological types analysed in the study. H&N, head and neck. b, Waterfall plot of cell death as assessed by propidium iodide staining (z score). Both non-cancer and cancer cells (n = 59 cell types) were transfected with four different siRNAs targeting NADSYN1 (red circles), and a non-targeting siNTC (open circle) was used a negative control. c, Heatmap illustrating cell death as measured by propidium iodide staining (z score). Different cancer cell types (n = 54 cell lines) were transfected with the indicated siRNAs. d, Intracellular measurement of NAD+/NADH (left) and NAD+ (right) levels in non-cancer (n = 5 cell types) and cancer (n = 21 cell types) cell lines. e, Intracellular measurement of NAD+/NADH (left) and NAD+ (right) levels in non-cancer cell lines (n = 5 cell types) and cancer cell lines amplified (PH-amp) or not amplified (non-PH-amp) for the PH-pathway enzymes NAPRT or NADSYN1 (n = 21 cell types). Box and whisker plots are as in Fig. 1d. Data are representative of independent biological replicates from three independent experiments. P values determined by one-way ANOVA with Tukey’s multiple comparisons test (b) or a two-tailed unpaired Student’s t-test (d, e).
Extended Data Fig. 4 Genetic depletion of genes encoding key enzymes of NAD biosynthesis pathways combined with metabolic addbacks identify mechanistic basis of NAD pathway addiction.
Cancer cell lines (n = 8 cell types) amplified for the PH-pathway enzymes NAPRT or NADSYN1 were transduced independently with the indicated DOX-inducible ishRNAs, followed by DOX treatment after puromycin selection. Non-cancer cell lines (black; n = 3 cell types) used as controls were also transduced with the indicated DOX-inducible ishRNAs. Cells were supplemented with fresh growth media and exogenous NAD+ (200 µM) or the indicated precursors, nicotinic acid (NA), nicotinic acid mononucleotide (NAMN), nicotinamide (NM), nicotinamide mononucleotide (NMN), nicotinamide riboside (NR), TRP or quinolinic acid (QA) at a concentration of 500 µM every 2–3 days. a, Representative images of colony formation assay using crystal violet staining from one of two independent experiments. Both biological replicates showed similar results. Cells stably expressing different ishRNAs were stained with crystal violet 15–18 days after transduction and selection. b, Heatmap illustrating absolute colony formation units. c, Immunoblotting for cleaved caspase-3 as a measure of cell death and to test for protein abundance for NAPRT and NAMPT in PH-amplified (left) and non-PH-amplified (right) cancer cells transduced with respective ishRNAs. Actin was used as a loading control. Representative blots from one of two independent experiments. Both biological replicates showed similar results. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 5 NAMPT enhancer drives NAD salvage-pathway addiction in cancer.
a, Luciferase enhancer reporter assay of the putative downstream enhancer. To test the effect of a predicted enhancer, the cis-regulatory region of the NAMPT locus was cloned into pGL3 reporter constructs in the direction indicated. Enhancer activity of the 4.641-kb cis-regulatory region corresponding to the H3K27ac and DHS peak was tested using a luciferase reporter assay, when present both upstream and downstream of the luciferase gene in a construct containing the NAMPT promoter. The pGL3 reporter plasmid containing the NAMPT promoter but without the enhancer region is used as a negative control (pGL3). Luciferase reporter assay measuring the enhancer activity (NAMPT-Enh) was tested in salvage-dependent, U87Sal-dep and HCT116Sal-dep cancer cells. Relative luciferase units are normalized to Renilla. b, NAMPT transcript levels as measured by qPCR (left), and intracellular measurement of NAD+ levels (right) in the indicated cells transduced with the KRAB-dCAS9 genetic repression system. c, Immunoblotting for abundance of cleaved caspase-3 in cells transduced with the KRAB-dCAS9 genetic repression system. Representative blots are from one of two independent experiments. Both biological replicates showed similar results. Actin was used as loading control. When quantifying NAD+ and cleaved caspase-3 abundance in H460Sal-dep, U87Sal-dep and HCT116Sal-dep cells, exogenous NAD+ (200 µM) was added to test for the rescue of the phenotype. See schematic in Fig. 3a for the design of KRAB-dCas9-mediated repression of the NAMPT enhancer embedded within the ‘B’ sub-region. Guide RNAs are as in Fig. 3a. d, Genome browser screenshot illustrating transcription factor (TF) ChIP–seq epigenome profiles across several Sal-dep cancer cells (HeLa, A549, K562 and SK-N-SH). The peach shaded region embedding the transcription factor ChIP–seq peaks indicates putative transcription factor recruitment sites that overlap NAMPT ‘B’ enhancer region (marked by a red square box at the bottom of the clustering, hg19 Chr7: 105,856,541–105,858,299). The grey shaded region corresponds to the NAMPT promoter (hg19_dna chr7:105,925,229–105,926,250). e, Transcript levels of MYC, MAX, STAT3, FOXM1 and GATA3 transcription factor genes in H460Sal-dep cancer cells after siRNA-mediated depletion. Bar plots are representative of five (a, b) and three (b, bottom right, e) independent biological replicates from independent experiments. Data are mean ± s.d. P values determined by one-way (a, b) or two-way (e) ANOVA with Tukey’s multiple comparisons test. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 6 In vivo demonstration of NAD metabolic pathway dependencies.
a, Schematic overview of experiment. OV4PH-amp cells stably expressing DOX-inducible shRNA against NAPRT, NADSYN1, NAMPT or NMRK1 were inoculated into the left flank of nude mice. H460Sal-dep cells stably expressing DOX-inducible shRNAs were inoculated into the right flank of the same nude mice. ishNTC was used as a non-targeting control for both tumour types. b, Tumour volume (left) and intratumoral NAD+ measurement (right) of nude mice bearing OV4PH-amp cell stably expressing DOX-inducible shRNA against NMRK1 taken at the end of experiment on day 30. Tumour volume was monitored over a 30-day period. DOX treatment was initiated on day 7 after implantation until the end of the experiment. c, e, g, Representative images illustrating TUNEL+ nuclei and Ki67+ cells from one of two independent experiments. Both biological replicates showed similar results. d, f, h, Quantification of TUNEL+ nuclei and Ki67+ cells. DAPI was used to stain DNA for TUNEL staining. For TUNEL+ nuclei, 10,000–12,000 cells were counted for each cohort; for Ki67+ cells, 15,000–20,000 cells were counted for each cohort. i, Immunoblotting for cleaved-caspase-3 as a measure of cell death and to test for protein abundance of NAMPT, NAPRT, NMRK1 and NADSYN1 in tumour tissues obtained from the indicated tumour types. Representative blots are from one of two independent experiments. Both biological replicates showed similar results. Actin was used as a loading control. Data are representative of eight (b, left, d, f, h) and five (b, right) independent biological replicates from two independent experiments. Data are mean tumour volume ± s.e.m. (n = 8 tumours per cohort), with values determined by two-way ANOVA on repeated measurements over time (b, left). Data in scatter plots are mean ± s.d., with P values determined by one-way ANOVA with Tukey’s multiple comparisons test (d, f, h). For gel source data, see Supplementary Fig. 1. ns, not significant.
Extended Data Fig. 7 In vivo genetic depletion of genes encoding key enzymes of NAD biosynthesis pathways combined with genetic rescue identify mechanistic basis of NAD pathway addiction.
a, Schematic overview of experiment. OV4PH-amp or H460Sal-dep cells stably expressing DOX-inducible shRNA targeting the 3′ UTR of the target genes were inoculated into the left flank of individual nude mice as indicated. The same clone of the stably engineered OV4PH-amp or H460Sal-dep cells but with expression of exogenous cDNA corresponding to the target not susceptible to silencing compared to the endogenous copy (ishNAPRT(+naprt-Flag)), (ishNAMPT(+nampt-Flag)) or (ishNAMPT+NMRK1(+nmrk1-Flag)) was inoculated into the right flank of individual mice as indicated. ishNTC was used as a non-targeting control inducible shRNA. b, Tumour volume from different tumour types as indicated. Tumour volume was monitored over a 30-day period. DOX treatment was initiated on day 7 after implantation until the end of the experiment. c, Intratumoral NAD+ measurement of nude mice bearing tumours taken at the end of experiment on day 30 for the indicated tumour types. d, Immunoblotting for NAPRT, NAMPT, NMRK1 and Flag in tumour tissues obtained from the indicated tumour types to check for protein abundance. Representative blots are from one of two independent experiments. Both biological replicates showed similar results. Actin was used as a loading control. Data are representative of eight (b) or six (c) independent biological replicates from two independent experiments. Data are mean tumour volume ± s.e.m. (n = 8 tumours per cohort), with P values determined by two-way ANOVA on repeated measurements over time (b). Data in scatter plots are mean ± s.d., with P values determined by one-way ANOVA with Tukey’s multiple comparisons test (c). For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 8 NAMPT deficiency leads to enzymatic bypass of the salvage pathway, successfully reprogramming NAD biosynthesis in cancer.
Genetically engineered salvage-dependent cancer cells including H460Sal-dep, HCT116Sal-dep and U87Sal-dep were transduced with two different shRNAs against NAMPT followed by puromycin selection. A non-targeting shNTC was used a control. a, Top, intracellular measurement of NAD+ levels. Middle, representative images of clonogenic survival assay using crystal violet staining from one of two independent experiments. Both biological replicates showed similar results. Bottom, quantification of colony formation units. Cells were stained with crystal violet 15–18 days after seeding. Salvage-dependent cancer cells with NAMPT stably silenced grown for an extended duration of time (long-term depletion) were later silenced with shRNA against NMRK1. b, Immunoblotting for cleaved caspase-3 as a measure of cell death and to test for protein abundance of NAMPT, NMRK1 and NAPRT. Representative blots are from one of two independent experiments. Both biological replicates showed similar results. Actin was used as a loading control. Salvage-dependent cancer cells stably silenced for NAMPT and grown for an extended duration of time (long-term depletion) were later silenced for NMRK1 using siRNA. c, d, Relative NMRK1 and NAMPT, NMRK2 or NAPRT transcript levels as measured by qPCR. Salvage-dependent cancer cells stably silenced for NAMPT and grown for an extended duration of time (long-term depletion), were later silenced with NMRK1 shRNA. e, Schematic overview of the model illustrating NAD pathway addiction in cancer is driven by two separate mechanisms—one that gets shaped by gene amplification (left), and the other through epigenetic reprogramming (right). The model demonstrates tissue context-based amplifications of genes encoding key enzymes (NAPRT and NADSYN1) of the PH-pathway and subsequent tumour cell dependence that is absolute and not subjected to enzymatic bypass rewiring. By contrast, epigenetically determined dependence on the NAMPT driven salvage-pathway is subject to enzymatic bypass, requiring combination therapies. In all panels, for short-term depletion, cells were seeded 7–10 days after transduction/selection, and for long-term depletion, cells were seeded ≥30 days after transduction/selection. Data are representative of five (a, top, c), three (d) and two (a, middle, bottom) independent biological replicates from independent experiments. Data in scatter plots are mean ± s.d., with P values determined by one-way ANOVA with Tukey’s multiple comparisons test (a, c, d). For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 9 Genetic depletion of NMRK1 in non-PH-amplified tumour cells enhances sensitivity to FK-866 inducing tumour cell death.
a, Heatmap illustrating cell death measured by propidium iodide staining (z score). Cancer cells were treated with increasing doses of FK-866 for 72 h. b, Intracellular measurement of NAD+ levels. c, Immunoblotting for cleaved caspase-3, in salvage-dependent cancer cells treated with 10 nM FK-866 for 72 h. Cells were supplemented with exogenous NAD+ (200 µM) or with the indicated precursors (NM, NMR, NR, NA) at a dose of 500 µM. d, Cell viability of non-cancer and cancer cells (PH-amp and sal-dep) stably silenced with shRNA against NMRK1, treated with increasing doses of FK-866 for 72 h. e, Schematic overview of experiment. OV4PH-amp (top) and H460Sal-dep (bottom) cells stably expressing shRNA against the target gene NMRK1 were implanted subcutaneously. f, Tumour volume of nude mice bearing stably engineered OV4PH-amp cells implanted subcutaneously. Tumour volume was monitored over a 24-day period. Mice were injected intraperitoneally with FK-866 twice daily. g, Intratumoral NAD+ measurement of nude mice bearing stably engineered OV4PH-amp tumours, taken at the end of experiment on day 24. h, Immunoblotting for cleaved caspase-3 as a measure of cell death and to test for protein abundance for NMRK1 in tumour tissues obtained from the indicated tumour types. Representative blots are from one of two independent experiments. Both biological replicates showed similar results. Actin was used as a loading control. Data are representative of three (b, d), eight (f) or six (g) independent biological replicates from independent experiments. Data in scatter plots (b, g) are mean ± s.d., with P values determined by one-way ANOVA with Tukey’s multiple comparisons test. For cell viability data (d), P values were determined by two-tailed unpaired Student’s t-test. Data in f are mean tumour volume ± s.e.m. (n = 8 tumours/cohort), with P values determined by two-way ANOVA on repeated measurements over time. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 10 Overexpression of rate-limiting NAD biosynthesis enzymes is not sufficient to generate or reverse metabolic addiction.
a, Immunoblotting for cleaved caspase-3 as a measure of cell-death and to test for abundance of NAMPT and NAPRT protein expression in non-cancer cells (IMR90, RPE-1) stably overexpressing NAMPT or NAPRT. Protein lysates from etoposide-treated H460 cancer cells was used as a control. b, Scattered plots represent percentage cell death as assessed by trypan blue exclusion assay. c, Intracellular measurement of NAD+ levels. d, e, Immunoblotting for cleaved caspase-3 as a measure of cell death and to test for abundance of NAPRT (d, in H460non-PH-amp) and NAMPT (e, in OV4PH-amp) protein expression. Protein lysates from etoposide-treated H460 or OV4 cancer cells was used as a control. f, Scattered plots represent percentage cell death as assessed by trypan blue exclusion assay. g, Intracellular measurement of NAD+ levels, after stable overexpression of NAPRT or NAMPT in H460 or in OV4 cancer cells as indicated. Stably engineered non-cancer and cancer cells (a–g) after selection were treated with FK-866 (10 nM) or NADSYN11i (2 μM) as indicated for 72 h. Representative blots are from one of two independent experiments. Both biological replicates showed similar results. Actin was used as a loading control (a, d, e). Data are representative of five (b, f) and three (c, g) independent biological replicates from independent experiments. Data are mean ± s.d. P values determined by two-way ANOVA with Tukey’s multiple comparisons test. For gel source data, see Supplementary Fig. 1.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-4
Supplementary Figure 1
This file contains Supplementary Figure 1, the uncropped western blot images
Source data
Rights and permissions
About this article
Cite this article
Chowdhry, S., Zanca, C., Rajkumar, U. et al. NAD metabolic dependency in cancer is shaped by gene amplification and enhancer remodelling. Nature 569, 570–575 (2019). https://doi.org/10.1038/s41586-019-1150-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1150-2
This article is cited by
-
Structural insights into Xanthomonas campestris pv. campestris NAD+ biosynthesis via the NAM salvage pathway
Communications Biology (2024)
-
Multi-omics analysis reveals NNMT as a master metabolic regulator of metastasis in esophageal squamous cell carcinoma
npj Precision Oncology (2024)
-
Dual-inhibition of NAMPT and PAK4 induces anti-tumor effects in 3D-spheroids model of platinum-resistant ovarian cancer
Cancer Gene Therapy (2024)
-
NAD pool as an antitumor target against cancer stem cells in head and neck cancer
Journal of Experimental & Clinical Cancer Research (2023)
-
DNMT3A mutation promotes leukemia development through NAM-NAD metabolic reprogramming
Journal of Translational Medicine (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.